New Chromosomal Abnormality Identified in Leukemia Associated with Down Syndrome
Study led by St. Jude Children's Research Hospital investigators expands understanding of acute lymphoblastic leukemia in children with Down syndrome, fueling hope for new treatment
Researchers identified a new chromosomal abnormality in acute lymphoblastic leukemia (ALL) that appears to work in concert with another mutation to give rise to cancer. This latest anomaly is particularly common in children with Down syndrome.
The findings have already resulted in new diagnostic tests and potential tools for tracking a patient's response to treatment. The research, led by scientists from St. Jude Children's Research Hospital, also highlights a new potential ALL treatment. Clinicians are already planning trials of an experimental medication targeting one of the altered genes.
"A substantial proportion of children with ALL lack one of the previously identified, common chromosomal abnormalities. Also, children with Down syndrome have an increased risk of ALL, but the reasons why are unclear," said Charles Mullighan, M.D., Ph.D., assistant member in the St. Jude Department of Pathology. Mullighan is senior author of the study, which involved scientists from 10 institutions in the U.S. and Italy. "Our results have provided important data regarding the mechanisms contributing to leukemia in these cases," he said.
Instead of the normal pairs of 23 chromosomes, individuals with Down syndrome inherit an extra copy of one chromosome, in this case chromosome 21. Chromosomes are made of DNA and carry the genes that serve as the assembly and operations manual for life. Down syndrome is associated with a variety of medical and developmental problems, including a 10-to-20--fold increased risk of ALL. But patients with Down syndrome rarely have the genetic and chromosomal alterations commonly associated with childhood ALL. Until recently the genetic basis of the elevated risk for these patients was unknown.
The new gene alteration was identified by St. Jude scientists following up on an earlier observation. They had previously found a recurring deletion in a region of DNA duplicated on the X and Y chromosomes. The region is known as pseudoautosomal region 1 or PAR1.
The PAR1 deletion was found only in patients with a subtype of ALL known as B-progenitor ALL. It was most common in children with both B-progenitor ALL and Down syndrome. In this study, investigators screened almost 400 children with ALL, including 75 patients with Down syndrome. The deletion was present in 7 percent of patients with B-progenitor ALL, but in more than half of the patients with both B progenitor and Down syndrome.
The deletion results in a fusion of two genes, P2RY8 and CRLF2. The fusion puts CRLF2 expression under the control of the P2RY8 promoter. As a result, CRLF2 expression jumps as much as 10 fold.
"CRLF2 over-expression identifies a group of ALL cases which were not previously well characterized, and suggests some novel treatment approaches that may improve patient survival. Patients with Down syndrome are particularly vulnerable to complications from standard chemotherapy, and could therefore benefit from novel therapies," said Karen Rabin, M.D., of Texas Children's Cancer Center and a study co-author. She is a Baylor College of Medicine assistant professor of pediatric hematology/oncology.
The CRLF2 protein normally forms part of a receptor where a small growth factor known as a cytokine binds to white blood cells known as lymphocytes. Both the cytokine, thymic stromal lymphopoietin (TSLP), and CRLF2 are known to play important roles in the development of immune cells known as T lymphocytes as well as in inflammation and allergic disease. They had not previously been linked to leukemia.
CRFL2 is the second gene implicated in development of B-progenitor ALL in patients with Down syndrome. The first, a gene called JAK2, was identified in 2008. JAK2 belongs to a family of genes that produce enzymes called kinases. If permanently switched on, kinases can trigger the uncontrolled cell growth that is a hallmark of cancer.
JAK mutations have also been linked to other cancers. Drugs targeting JAK kinases are already in clinical trials against a variety of blood disorders in adults. Additional trials are being planned against other subtypes of childhood ALL.
In this study, researchers reported a significant association between alterations in both the CRFL2 and JAK2 genes. Almost all JAK mutations were observed in patients with CRLF2 alterations. Almost 28 percent of children with Down syndrome and ALL had changes in both the CRFL2 and JAK genes.
"It has been a mystery as to why the JAK mutations in Down syndrome ALL are different from those seen in other cancers," Mullighan said. "Here we show that the JAK mutations in ALL are almost always observed together with a chromosomal alteration that results in over- expression of CRLF2."
When both the JAK mutation and increased CRLF2 production were introduced into white blood cells growing in the laboratory, those cells were transformed and no longer needed cytokines to grow. Neither genetic alteration on its own produced the same effect. Researchers also reported their impact could be weakened by the addition of drugs that target JAK mutations.
"We showed that the two proteins, CRLF2 and mutant JAK2, physically interact, and together transform white blood cells. This work has identified a new pathway contributing to the development of leukemia," Mullighan said. A next step is to determine if these mutations also interact in mouse models of ALL.
Other authors of this paper include J. Racquel Collins-Underwood, Letha A.A. Phillips, Wei Liu, Jing Ma, Elaine Coustan-Smith, Richard T. Williams, Jinjun Cheng, Ching-Hon Pui, Susana Raimondi and James Downing, all of St. Jude; Michael L. Loudin of Baylor; Jinghui Zhang of the National Cancer Institute (NCI); Richard Harvey and Cheryl Willman, of the University of New Mexico; Fady Mikhail and Andrew Carroll, of the University of Alabama at Birmingham; Nyla Heerema of Ohio State University; Giuseppe Basso, of the University of Padua, Padua, Italy; and Andrea Pession of the University of Bologna, Bologna, Italy.
The work was supported in part by National Cancer Institute, the Bear Necessities Pediatric Research Foundation, the Children's Cancer Research Foundation, the National Institutes of Health and ALSAC.
St. Jude Children's Research Hospital
St. Jude Children's Research Hospital is internationally recognized for its pioneering work in finding cures and saving children with cancer and other catastrophic diseases. Founded by late entertainer Danny Thomas and based in Memphis, Tenn., St. Jude freely shares its discoveries with scientific and medical communities around the world. No family ever pays for treatments not covered by insurance, and families without insurance are never asked to pay. St. Jude is financially supported by ALSAC, its fundraising organization. For more information, please visit www.stjude.org.
Sunday, August 23, 2009

Neutrinos are coming on Earth from Failed supernovae
spectacular supernovae (Fig. 1). The temperatures and pressures generated in these events are so intense they create a large burst of particles called neutrinos, which eventually reach Earth.
Now, Cecilia Lunardini at Arizona State University and RIKEN BNL Research Center in Upton, USA, has calculated that lots of neutrinos may also reach Earth from ‘failed supernovae’—huge stars that collapse without exploding to produce black holes1.
The neutrino contribution from these failed supernovae could greatly increase the total flux of neutrinos reaching Earth from millions of collapsing stars throughout the universe. Lunardini calls this total the ‘diffuse supernova neutrino flux’.
“In the diffuse flux, the contribution of each supernova is very small, but the total is detectable,” she says. “We only need to reach the right experimental sensitivity to start detecting it.”
Unfortunately, neutrinos are notoriously difficult to detect because they barely interact with other matter. One of the world’s best detectors is the Super-Kamiokande (‘Super-K’) neutrino observatory, situated in a mine beneath Gifu prefecture Japan, and even it requires 50,000 tons of ultra-pure water to scatter the neutrinos.
Lunardini decided to calculate whether a device like Super-K could detect neutrinos from supernovae collapsing into black holes.
“The idea that neutrinos are emitted in black-hole-forming collapses is not new,” she says. “The novelty of my work is in showing that these neutrinos can build up to a significant diffuse flux, thus adding to the flux from successful supernovae.”
In fact, Lunardini calculated that the Earth may receive up to one neutrino per square centimeter per second from failed supernovae. This is even more than the flux from successful supernovae, but probably beyond the detection limit of Super-K.
There is growing support in the scientific community to build larger, more sensitive neutrino detectors containing up to a million tons of water. Once these bigger detectors are built, Lunardini thinks it is only a matter of time before the diffuse neutrino flux can be measured. The results could reveal some fascinating new physics.
“[Failed supernovae] are very difficult to study with telescopes due to the fact that they do not explode but just disappear from the sky without much emission other than neutrinos,” says Lunardini. “The possibility to get information on these objects—even just to test their presence and how many there are in the universe—with neutrinos is exciting.”
Reference
1. Lunardini, C. Diffuse neutrino flux from failed supernovae. Physical Review Letters 102, 231101 (2009).
The corresponding author for this highlight is based at the RIKEN BNL Research Center Theory Group
Labels:
Failed supernovae,
Lunardini,
Neutrinos
Wednesday, August 19, 2009
Meningitis Bacteria penetrate the Blood-Brain Barrier
A specific protein on the surface of a common bacterial pathogen allows the bacteria to leave the bloodstream and enter the brain, initiating the deadly infection known as meningitis. The new finding, which may guide development of improved vaccines to protect those most vulnerable, including young infants and the elderly, is now available online in the Journal of Experimental Medicine.
"Streptococcus pneumoniae, commonly known as pneumococcus, is responsible for half the cases of bacterial meningitis in humans," said the study's senior author, Victor Nizet, MD, professor of pediatrics and pharmacy at the University of California, San Diego’s School of Medicine and Skaggs School of Pharmacy and Pharmaceutical Sciences. “As many as 30 percent of patients can die from this rapidly progressing infection, while half of survivors may be left with permanent neurological problems including deafness, seizures, intellectual deficits or motor disabilities.”
Meningitis develops when bacteria penetrate the "blood-brain barrier." Comprised of a single layer of highly specialized microvascular endothelial cells, the blood-brain barrier prevents most large molecules from entering into the cerebrospinal fluid, preserving an optimal biochemical environment for brain function.
The UC San Diego team investigated the functions of a protein known as NanA in order to discover how an entire bacterium can breech the blood-brain barrier and gain access to the central nervous system. NanA is produced by all strains of pneumococcus and displayed prominently on the bacteria's outer surface.
Through genetic manipulations, the researchers were able to remove the entire NanA protein, or just specific sections of the molecule, from the pathogen. They found that while normal pneumococci were able to bind, enter and penetrate through human brain microvascular endothelial cells, mutant bacteria lacking the NanA protein –or those expressing only a truncated version of the protein – largely lost these abilities. Conversely, when the full-length pneumococcal NanA protein was cloned and expressed on the surface of a nonpathogenic laboratory strain, the transformed bacteria gained the ability to bind and enter the same endothelial cells.
“Our tissue culture studies showed that the NanA protein was both necessary and sufficient for bacterial penetration of the blood brain barrier endothelial cells,” said Satoshi Uchiyama, MD, a postdoctoral fellow in the Nizet Laboratory and lead author on the study. “After infecting mice intravenously, we also found that far fewer NanA-deficient bacteria left the bloodstream and entered the brain, in comparison to mice infected with the normal pneumococcus.”
NanA is best known as an enzyme that cleaves and releases the sugar molecule known as sialic acid, which is present in abundance on the surface of all human cells. While this enzymatic activity played a small part in promoting NanA-mediated blood-brain barrier interactions, a much stronger role was identified for the outer tip of the protein. This tip seems to directly attach to the brain microvascular endothelial cells and then stimulate them to take in the pneumococcus.
“Antibodies directed against the NanA protein also strongly inhibited the ability of pneumococcus to attach to and invade the blood-brain barrier cells,” said Kelly Doran, PhD, an assistant professor at both UC San Diego School of Medicine and San Diego State University, who jointly supervised the project with Nizet.
Because NanA is expressed on the surface of all pneumococcal strains, it is an attractive candidate to include in a universal protein-based vaccine against pneumococcal infection according to Nizet, who is also on the medical staff of Rady Children's Hospital, San Diego. Currently, infants and the elderly are immunized with vaccines comprised of surface capsule sugars from 7 to 23 of the most common strains of pneumococcus.
“Our immune system generates antibodies more readily against protein rather than sugar vaccine antigens,” said Nizet. “Since at least 80 different pneumococcal capsule types exist, not all strains can be represented in the capsule sugar-based vaccines. An added benefit of an effective NanA vaccine would be to reduce the risk of pneumococcus spreading into the brain to cause meningitis.”
Ongoing research in the Nizet and Doran labs will seek to characterize the receptor on the blood-brain barrier cells to which NanA binds, and to explore whether similar processes contribute to the ability of other meningitis pathogens – such as group B streptococcus – to pass through the blood-brain barrier.
Additional contributors to the project were co-lead author Aaron Carlin, MD, PhD, Arya Khosravi, Shannon Weiman, Darin Quach, and George Hightower of the Department of Pediatrics at UC San Diego School of Medicine; Timothy Mitchell, PhD, of the University of Glasgow; and Anirban Banerjee, PhD, of the Department of Biology at San Diego State University. The research was supported by the National Institutes of Health, the American Heart Association, the Burroughs-Wellcome Fund and the Taylor Thomas Foundation.
A specific protein on the surface of a common bacterial pathogen allows the bacteria to leave the bloodstream and enter the brain, initiating the deadly infection known as meningitis. The new finding, which may guide development of improved vaccines to protect those most vulnerable, including young infants and the elderly, is now available online in the Journal of Experimental Medicine.
"Streptococcus pneumoniae, commonly known as pneumococcus, is responsible for half the cases of bacterial meningitis in humans," said the study's senior author, Victor Nizet, MD, professor of pediatrics and pharmacy at the University of California, San Diego’s School of Medicine and Skaggs School of Pharmacy and Pharmaceutical Sciences. “As many as 30 percent of patients can die from this rapidly progressing infection, while half of survivors may be left with permanent neurological problems including deafness, seizures, intellectual deficits or motor disabilities.”
Meningitis develops when bacteria penetrate the "blood-brain barrier." Comprised of a single layer of highly specialized microvascular endothelial cells, the blood-brain barrier prevents most large molecules from entering into the cerebrospinal fluid, preserving an optimal biochemical environment for brain function.
The UC San Diego team investigated the functions of a protein known as NanA in order to discover how an entire bacterium can breech the blood-brain barrier and gain access to the central nervous system. NanA is produced by all strains of pneumococcus and displayed prominently on the bacteria's outer surface.
Through genetic manipulations, the researchers were able to remove the entire NanA protein, or just specific sections of the molecule, from the pathogen. They found that while normal pneumococci were able to bind, enter and penetrate through human brain microvascular endothelial cells, mutant bacteria lacking the NanA protein –or those expressing only a truncated version of the protein – largely lost these abilities. Conversely, when the full-length pneumococcal NanA protein was cloned and expressed on the surface of a nonpathogenic laboratory strain, the transformed bacteria gained the ability to bind and enter the same endothelial cells.
“Our tissue culture studies showed that the NanA protein was both necessary and sufficient for bacterial penetration of the blood brain barrier endothelial cells,” said Satoshi Uchiyama, MD, a postdoctoral fellow in the Nizet Laboratory and lead author on the study. “After infecting mice intravenously, we also found that far fewer NanA-deficient bacteria left the bloodstream and entered the brain, in comparison to mice infected with the normal pneumococcus.”
NanA is best known as an enzyme that cleaves and releases the sugar molecule known as sialic acid, which is present in abundance on the surface of all human cells. While this enzymatic activity played a small part in promoting NanA-mediated blood-brain barrier interactions, a much stronger role was identified for the outer tip of the protein. This tip seems to directly attach to the brain microvascular endothelial cells and then stimulate them to take in the pneumococcus.
“Antibodies directed against the NanA protein also strongly inhibited the ability of pneumococcus to attach to and invade the blood-brain barrier cells,” said Kelly Doran, PhD, an assistant professor at both UC San Diego School of Medicine and San Diego State University, who jointly supervised the project with Nizet.
Because NanA is expressed on the surface of all pneumococcal strains, it is an attractive candidate to include in a universal protein-based vaccine against pneumococcal infection according to Nizet, who is also on the medical staff of Rady Children's Hospital, San Diego. Currently, infants and the elderly are immunized with vaccines comprised of surface capsule sugars from 7 to 23 of the most common strains of pneumococcus.
“Our immune system generates antibodies more readily against protein rather than sugar vaccine antigens,” said Nizet. “Since at least 80 different pneumococcal capsule types exist, not all strains can be represented in the capsule sugar-based vaccines. An added benefit of an effective NanA vaccine would be to reduce the risk of pneumococcus spreading into the brain to cause meningitis.”
Ongoing research in the Nizet and Doran labs will seek to characterize the receptor on the blood-brain barrier cells to which NanA binds, and to explore whether similar processes contribute to the ability of other meningitis pathogens – such as group B streptococcus – to pass through the blood-brain barrier.
Additional contributors to the project were co-lead author Aaron Carlin, MD, PhD, Arya Khosravi, Shannon Weiman, Darin Quach, and George Hightower of the Department of Pediatrics at UC San Diego School of Medicine; Timothy Mitchell, PhD, of the University of Glasgow; and Anirban Banerjee, PhD, of the Department of Biology at San Diego State University. The research was supported by the National Institutes of Health, the American Heart Association, the Burroughs-Wellcome Fund and the Taylor Thomas Foundation.
Labels:
blood-brain barrier,
Meningitis,
Meningitis bacteria
Friday, August 14, 2009

Oxytocin in Mammals and Mesotocin in Birds
What do flocks of birds have in common with trust, monogamy, and even breast milk? According to a new report in the journal Science, they are regulated by virtually identical neurochemicals in the brain, known as oxytocin in mammals and mesotocin in birds.
Neurobiologists at Indiana University showed that if the actions of mesotocin are blocked in the brains of zebra finches, a highly social songbird, the birds shift their social preferences. They spend significantly less time with familiar individuals and more time with unfamiliar individuals. The birds also become less social, preferring to spend less time with a large group of same-sex birds and more time with a smaller group. Conversely, if birds are administered mesotocin instead of the blocker, the finches become more social and prefer familiar partners.
Perhaps most striking is the fact that none of the treatments affect males -- only females.
According to James Goodson, lead author on the study, the sex differences in birds provide important clues to the evolutionary history of oxytocin functions in humans and other mammals. "Oxytocin is an evolutionarily descendant of mesotocin and has long been associated with female reproductive functions -- things such as pair bonding with males, giving birth, providing maternal care and ejecting milk for infants," said Goodson.
Goodson and colleagues have found hints of similar processes in fish, and he speculates that oxytocin-like neuropeptides have played special roles in female affiliation ever since the peptides first evolved. That was sometime around 450 million years ago, about the same time that jaws evolved.
"The ancient properties of this system appear to be retained in all major vertebrate groups, and date back to our common ancestor with sharks," says co-author Marcy Kingsbury, associate scientist at IU Bloomington.
But if all vertebrates possess similar neuropeptide circuits, why don't they all live in big groups -- flocks, schools or herds? A possible answer to that question is provided in the second part of the Science study. The authors speculated that the behavioral actions of mesotocin may differ across species depending upon the distribution of "receptors" for the chemical in the brain -- that is, places where mesotocin can attach to brain cells and alter their activity.
Using a radioactive compound that attaches to oxytocin-like receptors, the authors mapped the distribution of receptors in three finch species that form flocks and two species that are territorial and highly aggressive. What they found was that the flocking species had many more receptors in a part of the brain known as the lateral septum. And when they blocked those receptors in female zebra finches, the birds became less social.
According to Goodson, these findings suggest that it is actually the concentration and location of receptors that determines whether an individual prefers spending time in large groups. Natural selection could act to increase the number of receptors expressed by certain lateral septum neurons, or by altering the regions where receptor genes are expressed, depending on whether female sociality is favored or not among the individuals of a species.
If Goodson's discovery holds true for other birds and even mammals, the concentration of receptors for mesotocin (and oxytocin) in the lateral septum could accurately predict whether an individual is naturally gregarious.
"The lateral septum is structurally very similar in reptiles, birds and mammals," Goodson said. "To our knowledge, it plays an important role in the social and reproductive behaviors of all land vertebrates."
What might be next for Goodson's research group?
"We still don't understand why mesotocin and oxytocin are so potent in females, but not always in males," Goodson said. "And we also don't fully understand how the lateral septum functions to influence sociality." But he is convinced that his group's ongoing studies of songbirds will soon provide the answers.
IU Bloomington Associate Scientist Marcy Kingsbury, postdoctoral fellow David Kabelik, research associate Sara Schrock and Ph.D. student James Klatt also contributed to this research. It was funded with a grant from the National Institutes of Health (NIMH).
Friday, August 07, 2009
Researchers Make Stem Cells from Developing Sperm
The promise of stem cell therapy may lie in uncovering how adult cells revert back into a primordial, stem cell state, whose fate is yet to be determined. Now, cell scientists at the Johns Hopkins University School of Medicine have identified key molecular players responsible for this reversion in fruit fly sperm cells. Reporting online this week in Cell Stem Cell, researchers show that two proteins are responsible redirecting cells on the way to becoming sperm back to stem cells.
“We knew from our previous work that cells destined to be sperm could revert back to being stem cells, but we didn't know how,” says Erika Matunis, Ph.D., an associate professor of cell biology at the Johns Hopkins University School of Medicine. “Since, dedifferentiation is an interesting phenomenon probably occurring in a lot of different stem cell populations, we wanted to know more about the process.“
Like all stem cells, each of the nine stem cells in the fly testis divides to form two daughter cells: One stays a stem cell and the other differentiates into an adult cell, in this case, a sperm cell. To figure out what might cause sperm cells to revert or dedifferentiate, Matunis’s research team genetically altered the flies so that both cells become sperm, reducing the stem cell population in the testis to nothing.
About a week later, the team examined these fly testes and found that the stem cells had been repopulated.
To figure out how this was happening, the researchers first suspected two proteins—Jak and STAT—known to act together to help stem cells maintain their stem cell-ness. The team genetically altered flies to reduce the activity of Jak and STAT in the testis. Counting the number of cells, they found that the loss of Jak-STAT caused fewer cells to revert to stem cells; only 60 percent of testes regained stem cells compared to 97 percent in normal Jak-STAT-containing testes.
“We now know that in the fly testis, interfering with Jak-STAT signaling interferes with the process of dedifferentiation,” says Matunis.
Next, Matunis would like to figure out how Jak and STAT control dedifferentiation. “We don't know if a cell is just reversing all of the steps to go back to being a stem cell or if it is doing something totally new and different, but we’re eager to find out,” she says.
The promise of stem cell therapy may lie in uncovering how adult cells revert back into a primordial, stem cell state, whose fate is yet to be determined. Now, cell scientists at the Johns Hopkins University School of Medicine have identified key molecular players responsible for this reversion in fruit fly sperm cells. Reporting online this week in Cell Stem Cell, researchers show that two proteins are responsible redirecting cells on the way to becoming sperm back to stem cells.
“We knew from our previous work that cells destined to be sperm could revert back to being stem cells, but we didn't know how,” says Erika Matunis, Ph.D., an associate professor of cell biology at the Johns Hopkins University School of Medicine. “Since, dedifferentiation is an interesting phenomenon probably occurring in a lot of different stem cell populations, we wanted to know more about the process.“
Like all stem cells, each of the nine stem cells in the fly testis divides to form two daughter cells: One stays a stem cell and the other differentiates into an adult cell, in this case, a sperm cell. To figure out what might cause sperm cells to revert or dedifferentiate, Matunis’s research team genetically altered the flies so that both cells become sperm, reducing the stem cell population in the testis to nothing.
About a week later, the team examined these fly testes and found that the stem cells had been repopulated.
To figure out how this was happening, the researchers first suspected two proteins—Jak and STAT—known to act together to help stem cells maintain their stem cell-ness. The team genetically altered flies to reduce the activity of Jak and STAT in the testis. Counting the number of cells, they found that the loss of Jak-STAT caused fewer cells to revert to stem cells; only 60 percent of testes regained stem cells compared to 97 percent in normal Jak-STAT-containing testes.
“We now know that in the fly testis, interfering with Jak-STAT signaling interferes with the process of dedifferentiation,” says Matunis.
Next, Matunis would like to figure out how Jak and STAT control dedifferentiation. “We don't know if a cell is just reversing all of the steps to go back to being a stem cell or if it is doing something totally new and different, but we’re eager to find out,” she says.
Plastics That Convert Light to Electricity Could Have a Big impact
Researchers the world over are striving to develop organic solar cells that can be produced easily and inexpensively as thin films that could be widely used to generate electricity.
But a major obstacle is coaxing these carbon-based materials to reliably form the proper structure at the nanoscale (tinier than 2-millionths of an inch) to be highly efficient in converting light to electricity. The goal is to develop cells made from low-cost plastics that will transform at least 10 percent of the sunlight that they absorb into usable electricity and can be easily manufactured.
A research team headed by David Ginger, a University of Washington associate professor of chemistry, has found a way to make images of tiny bubbles and channels, roughly 10,000 times smaller than a human hair, inside plastic solar cells. These bubbles and channels form within the polymers as they are being created in a baking process, called annealing, that is used to improve the materials' performance.
The researchers are able to measure directly how much current each tiny bubble and channel carries, thus developing an understanding of exactly how a solar cell converts light into electricity. Ginger believes that will lead to a better understanding of which materials created under which conditions are most likely to meet the 10 percent efficiency goal.
As researchers approach that threshold, nanostructured plastic solar cells could be put into use on a broad scale, he said. As a start, they could be incorporated into purses or backpacks to charge cellular phones or mp3 players, but eventually they could make in important contribution to the electrical power supply.
Most researchers make plastic solar cells by blending two materials together in a thin film, then baking them to improve their performance. In the process, bubbles and channels form much as they would in a cake batter. The bubbles and channels affect how well the cell converts light into electricity and how much of the electric current actually gets to the wires leading out of the cell. The number of bubbles and channels and their configuration can be altered by how much heat is applied and for how long.
The exact structure of the bubbles and channels is critical to the solar cell's performance, but the relationship between baking time, bubble size, channel connectivity and efficiency has been difficult to understand. Some models used to guide development of plastic solar cells even ignore the structure issues and assume that blending the two materials into a film for solar cells will produce a smooth and uniform substance. That assumption can make it difficult to understand just how much efficiency can be engineered into a polymer, Ginger said.
For the current research, the scientists worked with a blend of polythiophene and fullerene, model materials considered basic to organic solar cell research because their response to forces such as heating can be readily extrapolated to other materials. The materials were baked together at different temperatures and for different lengths of time.
Ginger is the lead author of a paper documenting the work, published online last month by the American Chemical Society journal Nano Letters and scheduled for a future print edition. Coauthors are Liam Pingree and Obadiah Reid of the UW. The research was funded by the National Science Foundation and the U.S. Department of Energy.
Ginger noted that the polymer tested is not likely to reach the 10 percent efficiency threshold. But the results, he said, will be a useful guide to show which new combinations of materials and at what baking time and temperature could form bubbles and channels in a way that the resulting polymer might meet the standard.
Such testing can be accomplished using a very small tool called an atomic force microscope, which uses a needle similar to the one that plays records on an old-style phonograph to make a nanoscale image of the solar cell. The microscope, developed in Ginger's lab to record photocurrent, comes to a point just 10 to 20 nanometers across (a human hair is about 60,000 nanometers wide). The tip is coated with platinum or gold to conduct electrical current, and it traces back and forth across the solar cell to record the properties.
As the microscope traces back and forth over a solar cell, it records the channels and bubbles that were created as the material was formed. Using the microscope in conjunction with the knowledge gained from the current research, Ginger said, can help scientists determine quickly whether polymers they are working with are ever likely to reach the 10 percent efficiency threshold.
Making solar cells more efficient is crucial to making them cost effective, he said. And if costs can be brought low enough, solar cells could offset the need for more coal-generated electricity in years to come.
"The solution to the energy problem is going to be a mix, but in the long term solar power is going to be the biggest part of that mix," he said.
Researchers the world over are striving to develop organic solar cells that can be produced easily and inexpensively as thin films that could be widely used to generate electricity.
But a major obstacle is coaxing these carbon-based materials to reliably form the proper structure at the nanoscale (tinier than 2-millionths of an inch) to be highly efficient in converting light to electricity. The goal is to develop cells made from low-cost plastics that will transform at least 10 percent of the sunlight that they absorb into usable electricity and can be easily manufactured.
A research team headed by David Ginger, a University of Washington associate professor of chemistry, has found a way to make images of tiny bubbles and channels, roughly 10,000 times smaller than a human hair, inside plastic solar cells. These bubbles and channels form within the polymers as they are being created in a baking process, called annealing, that is used to improve the materials' performance.
The researchers are able to measure directly how much current each tiny bubble and channel carries, thus developing an understanding of exactly how a solar cell converts light into electricity. Ginger believes that will lead to a better understanding of which materials created under which conditions are most likely to meet the 10 percent efficiency goal.
As researchers approach that threshold, nanostructured plastic solar cells could be put into use on a broad scale, he said. As a start, they could be incorporated into purses or backpacks to charge cellular phones or mp3 players, but eventually they could make in important contribution to the electrical power supply.
Most researchers make plastic solar cells by blending two materials together in a thin film, then baking them to improve their performance. In the process, bubbles and channels form much as they would in a cake batter. The bubbles and channels affect how well the cell converts light into electricity and how much of the electric current actually gets to the wires leading out of the cell. The number of bubbles and channels and their configuration can be altered by how much heat is applied and for how long.
The exact structure of the bubbles and channels is critical to the solar cell's performance, but the relationship between baking time, bubble size, channel connectivity and efficiency has been difficult to understand. Some models used to guide development of plastic solar cells even ignore the structure issues and assume that blending the two materials into a film for solar cells will produce a smooth and uniform substance. That assumption can make it difficult to understand just how much efficiency can be engineered into a polymer, Ginger said.
For the current research, the scientists worked with a blend of polythiophene and fullerene, model materials considered basic to organic solar cell research because their response to forces such as heating can be readily extrapolated to other materials. The materials were baked together at different temperatures and for different lengths of time.
Ginger is the lead author of a paper documenting the work, published online last month by the American Chemical Society journal Nano Letters and scheduled for a future print edition. Coauthors are Liam Pingree and Obadiah Reid of the UW. The research was funded by the National Science Foundation and the U.S. Department of Energy.
Ginger noted that the polymer tested is not likely to reach the 10 percent efficiency threshold. But the results, he said, will be a useful guide to show which new combinations of materials and at what baking time and temperature could form bubbles and channels in a way that the resulting polymer might meet the standard.
Such testing can be accomplished using a very small tool called an atomic force microscope, which uses a needle similar to the one that plays records on an old-style phonograph to make a nanoscale image of the solar cell. The microscope, developed in Ginger's lab to record photocurrent, comes to a point just 10 to 20 nanometers across (a human hair is about 60,000 nanometers wide). The tip is coated with platinum or gold to conduct electrical current, and it traces back and forth across the solar cell to record the properties.
As the microscope traces back and forth over a solar cell, it records the channels and bubbles that were created as the material was formed. Using the microscope in conjunction with the knowledge gained from the current research, Ginger said, can help scientists determine quickly whether polymers they are working with are ever likely to reach the 10 percent efficiency threshold.
Making solar cells more efficient is crucial to making them cost effective, he said. And if costs can be brought low enough, solar cells could offset the need for more coal-generated electricity in years to come.
"The solution to the energy problem is going to be a mix, but in the long term solar power is going to be the biggest part of that mix," he said.
Monday, August 03, 2009

Breaking News: Major breakthrough in organ replacement regenerative therapies
Research group headed by Takashi Tsuji demonstrates in regenerating
bioengineered “fully functional organ (tooth)”
Substantial advance in the development of next-generationA research group led by Takashi Tsuji (Professor in the Research Institute for Science
and Technology, Tokyo University of Science, and Director of Organ Technologies Inc.) has
demonstrated in growing new organs in adult mice. Tsuji is a research team member in
“Health Labor Sciences Research Grant: Research on Regenerative Medicine for Clinical
Application (Domain Leader: Professor Akira Yamaguchi of Tokyo Medical and Dental
University)”, and “Priority Domain Research: Bio-engineering (Domain Leader: Professor
Toshio Fukuda of Nagoya University)”. In transplantation experiments using the tooth as a
model, a bioengineered tooth germ develops into a fully functioning bioengineered tooth with
sufficient hardness for mastication and a functional responsiveness to mechanical stress in the
maxillofacial region. The research also provided the results that the nerve fibers that have
re-entered the pulp and periodontal ligament (PDL) tissues of the bioengineered tooth have
proper perceptive potential in response to noxious stimulations such as orthodontic treatment
and pulp stimulation.
This research is expected to substantially advance in the development of “tooth
regenerative therapy”, which have potential as next-generation regenerative therapies for
replacing diseased or damaged teeth with bioengineered teeth. Specifically it will not only
promote “tooth regenerative therapy”, whereby organ germs of bioengineered teeth are
transplanted into the jaw bone to grow “3rd generation tooth”, but is expected to evolve into a
wide variety of organ regenerative technologies for liver, kidney and other organs.
This research outcome was the fruit of joint research with Professor Teruko
Takano-Yamamoto (Division of Orthodontics and Dentofacial Orthopedics, Graduate School
of Dentistry, Tohoku University, Japan) and Professor Shohei Kasugai (Oral and Maxillofacial
Surgery, Department of Oral Restitution, Division of Oral Health Sciences, Graduate School,
Tokyo Medical and Dental University, Japan). It was announced in an Advance Online
Publication of the US scientific journal “Proc. Natl. Acad. Sci. USA.”
“organ replacement regenerative therapies”
Wednesday, July 15, 2009
New Tools For Discovering DNA Variations In Crop Genomes
The study of human genetics has been a successful venture for researchers in recent years. Several million single-nucleotide polymorphisms (SNPs) have been identified from the whole-genome resequencing of multiple individuals, which have served as genetic markers to pinpoint genes controlling common human diseases. In contrast, the genome of a single cultivar or line has yet to be sequenced in its entirety for most crops of economic or societal importance. This slow pace of genomic progress can be mostly explained by the high costs and technical difficulties associated with sequencing crop genomes, which tend to be large in size and complex—containing a high amount of repetitive DNA and duplicated genes that are highly similar in sequence.
With the advent of high-throughput DNA sequencing technologies, it is now possible to cheaply and rapidly sequence hundreds of millions of bases in a matter of hours. A team of scientists at Cornell University (Ithaca, NY), the United States Department of Agriculture-Agriculture Research Service (USDA-ARS), Cold Spring Harbor Laboratory (Cold Spring Harbor, NY), Roche Applied Science Corp. (Indianapolis, IN) and 454 Life Sciences (Branford, CT), have developed molecular and computational tools for the efficient and accurate identification of gene-enriched SNPs in crops. The large, complex genome of maize was used to evaluate these tools.
The study was funded by the National Science Foundation (NSF), Roche Applied Science Corp., and the USDA-ARS. Results from the study were published in the July 2009 issue of The Plant Genome.
In this research collaboration, an existing molecular technique was modified to enable gene-enrichment and resequencing of maize inbred lines B73 and Mo17 with massively parallel pyrosequencing. In addition, a custom computational pipeline was developed to analyze and assemble short reads, identify correctly mapped reads, and call high quality SNPs. With the implementation of these methods, the authors identified 126,683 gene-enriched SNPs between B73 and Mo17 at high accuracy.
“Next-generation sequencing technologies will greatly accelerate the resequencing of multiple to numerous individuals for every major crop species,” says Michael Gore, first co-author of the study. “Such efforts will facilitate the construction of SNP datasets on the order of millions that can be used in whole-genome association studies to assess the contribution of SNPs—common or rare—to complex traits. What we have learned from this pilot study will help us to construct a community SNP resource in maize that is comparable in scale to that of the human haplotype map”.
Although the majority of SNPs do not contribute to phenotypic variation, plant breeders and geneticists alike are interested in using SNPs as genetic markers. As a genetic marker, SNPs can be used for studies of genetic diversity and in the selection of superior plants. The SNPs identified in this study can be used for high-resolution genetic mapping of agronomic traits, which could eventually lead to the development of improved commercial maize hybrids.
The study of human genetics has been a successful venture for researchers in recent years. Several million single-nucleotide polymorphisms (SNPs) have been identified from the whole-genome resequencing of multiple individuals, which have served as genetic markers to pinpoint genes controlling common human diseases. In contrast, the genome of a single cultivar or line has yet to be sequenced in its entirety for most crops of economic or societal importance. This slow pace of genomic progress can be mostly explained by the high costs and technical difficulties associated with sequencing crop genomes, which tend to be large in size and complex—containing a high amount of repetitive DNA and duplicated genes that are highly similar in sequence.
With the advent of high-throughput DNA sequencing technologies, it is now possible to cheaply and rapidly sequence hundreds of millions of bases in a matter of hours. A team of scientists at Cornell University (Ithaca, NY), the United States Department of Agriculture-Agriculture Research Service (USDA-ARS), Cold Spring Harbor Laboratory (Cold Spring Harbor, NY), Roche Applied Science Corp. (Indianapolis, IN) and 454 Life Sciences (Branford, CT), have developed molecular and computational tools for the efficient and accurate identification of gene-enriched SNPs in crops. The large, complex genome of maize was used to evaluate these tools.
The study was funded by the National Science Foundation (NSF), Roche Applied Science Corp., and the USDA-ARS. Results from the study were published in the July 2009 issue of The Plant Genome.
In this research collaboration, an existing molecular technique was modified to enable gene-enrichment and resequencing of maize inbred lines B73 and Mo17 with massively parallel pyrosequencing. In addition, a custom computational pipeline was developed to analyze and assemble short reads, identify correctly mapped reads, and call high quality SNPs. With the implementation of these methods, the authors identified 126,683 gene-enriched SNPs between B73 and Mo17 at high accuracy.
“Next-generation sequencing technologies will greatly accelerate the resequencing of multiple to numerous individuals for every major crop species,” says Michael Gore, first co-author of the study. “Such efforts will facilitate the construction of SNP datasets on the order of millions that can be used in whole-genome association studies to assess the contribution of SNPs—common or rare—to complex traits. What we have learned from this pilot study will help us to construct a community SNP resource in maize that is comparable in scale to that of the human haplotype map”.
Although the majority of SNPs do not contribute to phenotypic variation, plant breeders and geneticists alike are interested in using SNPs as genetic markers. As a genetic marker, SNPs can be used for studies of genetic diversity and in the selection of superior plants. The SNPs identified in this study can be used for high-resolution genetic mapping of agronomic traits, which could eventually lead to the development of improved commercial maize hybrids.
Wood Burning Stoves have been Reevaluated
The stress of rising natural gas prices is leading many consumers to rethink how they heat their homes. For some this means moving towards modern alternative energy options, while others have been turning to a more traditional method for a solution to these rising costs. In Canada and the United States, wood burning stoves have been reevaluated as a potentially viable option for home heating.
The case for modern woodstoves has developed with the improvement of the products on the market, as wood heating technology has substantially advanced in recent years. With the advanced secondary combustion systems on Environmental Protection Agency certified woodstoves, they are now 95% more efficient than their predecessors.
Dr. Paul Grogan, a plant and ecosystem ecologist and Canadian Research Chair (II) at Queen’s University in Kingston, Ontario conducted a case study on the benefits of woodstoves with the help of final-year undergraduate and first year graduate students. He determined that adding a woodstove to the home can help both consumers heating costs as well as the environment.
The environmental sustainability of woodstove use is dependent upon the consumption of wood from sustainably managed woodlots, as the carbon released is reused as the next generation of trees grows. Annual gross CO2 emissions did in fact increase from 12,610 kg (i.e., ~2.5 metric tons CO2/person per year) to 17,330 kg after the installation of the wood stove. But while this gross amount did increase, the net carbon released by the combustion is negligible, the only surplus coming from the harvest and transport. Based on an average growing time of 130 years before harvest for local Ontario tree species, a woodlot or forest 3.5 hectares in size would provide an indefinite supply of wood heat for a household without a net increase in carbon emissions.
In the case study, adding a woodstove to the ground floor of a 3200ft2 home reduced the mean annual gas cost by 60%; from $2260 to $880. The annual cost of the wood fuel for the woodstove amounted to $1330 for 5 full cords (a cord is 8 feet long by 4 feet high by 4 feet wide - 128ft3 ). This was a yearly savings of $50 at market fossil fuel prices of 2005-2007 without taking into account rising fossil fuel prices or the impending carbon tax. Should these variables come into play Dr. Grogan estimated that the domestic heating costs would be reduced by 25%. This translates into a potential savings of $920 in the first 3 years.
The stress of rising natural gas prices is leading many consumers to rethink how they heat their homes. For some this means moving towards modern alternative energy options, while others have been turning to a more traditional method for a solution to these rising costs. In Canada and the United States, wood burning stoves have been reevaluated as a potentially viable option for home heating.
The case for modern woodstoves has developed with the improvement of the products on the market, as wood heating technology has substantially advanced in recent years. With the advanced secondary combustion systems on Environmental Protection Agency certified woodstoves, they are now 95% more efficient than their predecessors.
Dr. Paul Grogan, a plant and ecosystem ecologist and Canadian Research Chair (II) at Queen’s University in Kingston, Ontario conducted a case study on the benefits of woodstoves with the help of final-year undergraduate and first year graduate students. He determined that adding a woodstove to the home can help both consumers heating costs as well as the environment.
The environmental sustainability of woodstove use is dependent upon the consumption of wood from sustainably managed woodlots, as the carbon released is reused as the next generation of trees grows. Annual gross CO2 emissions did in fact increase from 12,610 kg (i.e., ~2.5 metric tons CO2/person per year) to 17,330 kg after the installation of the wood stove. But while this gross amount did increase, the net carbon released by the combustion is negligible, the only surplus coming from the harvest and transport. Based on an average growing time of 130 years before harvest for local Ontario tree species, a woodlot or forest 3.5 hectares in size would provide an indefinite supply of wood heat for a household without a net increase in carbon emissions.
In the case study, adding a woodstove to the ground floor of a 3200ft2 home reduced the mean annual gas cost by 60%; from $2260 to $880. The annual cost of the wood fuel for the woodstove amounted to $1330 for 5 full cords (a cord is 8 feet long by 4 feet high by 4 feet wide - 128ft3 ). This was a yearly savings of $50 at market fossil fuel prices of 2005-2007 without taking into account rising fossil fuel prices or the impending carbon tax. Should these variables come into play Dr. Grogan estimated that the domestic heating costs would be reduced by 25%. This translates into a potential savings of $920 in the first 3 years.
Wednesday, July 01, 2009
First DNA-based Reconstruction of the Giant Extinct Moa Bird
Scientists have performed the first DNA-based reconstruction of the giant extinct moa bird, using prehistoric feathers recovered from caves and rock shelters in New Zealand.
Researchers from the University of Adelaide and Landcare Research in New Zealand have identified four different moa species after retrieving ancient DNA from moa feathers believed to be at least 2500 years old.
The giant birds – measuring up to 2.5 metres and weighing 250 kilograms – were the dominant animals in New Zealand’s pre-human environment but were quickly exterminated after the arrival of the Maori around 1280AD.
PhD student Nicolas Rawlence from the University’s Australian Centre for Ancient DNA says until now, the scientific community has not known what the 10 different species of moa looked like. ”By using ancient DNA we have been able to connect feathers to four different moa species,” he says.
The researchers compared the feathers to others found in the sediments from red-crowned parakeets that are still living today, determining they had not faded or changed in colour. They then reconstructed the appearance of the stout-legged moa, heavy-footed moa, upland moa and the South Island giant moa.
Their findings were published today in the Proceedings of the Royal Society of London Series B.
“The surprising thing is that while many of the species had a similar, relatively plain brown plumage for camouflage, some had white-tipped feathers to create a speckled appearance,” Mr Rawlence says.
A co-author of the study, Dr Jamie Wood from Landcare Research, says it is likely that the drab colouring was driven by selection to avoid predation by the extinct Haast’s eagle, the largest and most powerful eagle in the world.
The research team also demonstrated that it is possible to retrieve DNA from all parts of the ancient feathers, not just the tip of the quill, as previously thought.
“This important finding opens the way to study DNA from museum bird skins while causing almost no damage to these valuable specimens, just by clipping a small part of a single feather,” says Dr Kyle Armstrong from the Australian Centre for Ancient DNA (ACAD).
ACAD Director Professor Alan Cooper says this finding suggests it may be possible to reconstruct the appearance of other extinct birds using feathers from fossil deposits.
“There are so many enigmatic extinct species that it would be great to see ‘clothed’," Professor Cooper says.
Scientists have performed the first DNA-based reconstruction of the giant extinct moa bird, using prehistoric feathers recovered from caves and rock shelters in New Zealand.
Researchers from the University of Adelaide and Landcare Research in New Zealand have identified four different moa species after retrieving ancient DNA from moa feathers believed to be at least 2500 years old.
The giant birds – measuring up to 2.5 metres and weighing 250 kilograms – were the dominant animals in New Zealand’s pre-human environment but were quickly exterminated after the arrival of the Maori around 1280AD.
PhD student Nicolas Rawlence from the University’s Australian Centre for Ancient DNA says until now, the scientific community has not known what the 10 different species of moa looked like. ”By using ancient DNA we have been able to connect feathers to four different moa species,” he says.
The researchers compared the feathers to others found in the sediments from red-crowned parakeets that are still living today, determining they had not faded or changed in colour. They then reconstructed the appearance of the stout-legged moa, heavy-footed moa, upland moa and the South Island giant moa.
Their findings were published today in the Proceedings of the Royal Society of London Series B.
“The surprising thing is that while many of the species had a similar, relatively plain brown plumage for camouflage, some had white-tipped feathers to create a speckled appearance,” Mr Rawlence says.
A co-author of the study, Dr Jamie Wood from Landcare Research, says it is likely that the drab colouring was driven by selection to avoid predation by the extinct Haast’s eagle, the largest and most powerful eagle in the world.
The research team also demonstrated that it is possible to retrieve DNA from all parts of the ancient feathers, not just the tip of the quill, as previously thought.
“This important finding opens the way to study DNA from museum bird skins while causing almost no damage to these valuable specimens, just by clipping a small part of a single feather,” says Dr Kyle Armstrong from the Australian Centre for Ancient DNA (ACAD).
ACAD Director Professor Alan Cooper says this finding suggests it may be possible to reconstruct the appearance of other extinct birds using feathers from fossil deposits.
“There are so many enigmatic extinct species that it would be great to see ‘clothed’," Professor Cooper says.
Wednesday, June 24, 2009

First Direct Visualization of Memory Formation in the Brain
FINDINGS: UCLA and McGill University researchers have, for the first time, “photographed” a memory in the making. The study clarifies one of the ways in which connections in the brain between nerve cells, called synapses, can be changed with experience. The phenomenon is called “synaptic plasticity,” and is the foundation for how we learn and remember. As we learn, the memories are stored in changes in the strength and/or number of synaptic connections between nerve cells in our brain. Long lasting changes in synaptic connections are required for long-term memories, and the persistence of these changes requires new gene expression. This is the first study to use fluorescent imaging to directly visualize protein synthesis at individual synapses during learning related synaptic plasticity.
IMPACT: Understanding how synapses can change with experience is critical to understanding behavioral plasticity, and to understanding diseases in which learning and experience-dependent behaviors are impaired. Such diseases include mental retardation, Alzheimer’s disease, as well as anxiety and mood disorders. It also can elucidate potential strategies for improving normal cognition and behavioral plasticity.
JOURNAL: The research appears in the June 19 edition of the journal Science.
AUTHORS: Senior author Kelsey Martin, associate professor of psychiatry and biological chemistry; Dan Ohtan Wang, Sang Mok Kim, Yali Zhao, Hongik Hwang, Satoru K. Miura, all of UCLA; and Wayne S. Sossin, McGill University.
HOW: The researchers used sensory and motor neurons from the sea slug Aplysia Californica that can form connections in culture. The neurons were stimulated with serotonin, which strengthens the synapses, and allowed them to detect new protein synthesis—the making of a memory— using a “translational reporter,” a fluorescent protein that can be easily detected and tracked.
MORE: This is the first study to directly visualize protein synthesis at individual synapses during a long-lasting form of synaptic plasticity. The studies revealed an exquisite level of control over the specificity of regulation of new protein synthesis. “While this was not really surprising to us given the complexity of information processing in the brain,” said Martin, “visualizing the process of protein synthesis at individual synapses, and beginning to discern the elegance of its regulation, leaves us, as biologists, with a wonderful sense of awe.”
Funding: This study was funded by the National Institutes of Health, the WM Keck Foundation, and the Canadian Institutes of Health Research. The authors report no conflict of interest.
Friday, June 19, 2009
Bacteria Can Plan Ahead
Bacteria can anticipate a future event and prepare for it, according to new research at the Weizmann Institute of Science. In a paper that appeared in the June 17, 2009 issue of Nature, Prof. Yitzhak Pilpel, doctoral student Amir Mitchell, and research associate Dr. Orna Dahan of the Institute’s Molecular Genetics Department, together with Prof. Martin Kupiec and Gal Romano of Tel Aviv University, examined microorganisms living in environments that change in predictable ways. Their findings show that these microorganisms’ genetic networks are hard-wired to “foresee” what comes next in the sequence of events and begin responding to the new state of affairs before its onset.
E. coli bacteria, for instance, which normally cruise harmlessly down the digestive tract, encounter a number of different environments on their way. In particular, they find that one type of sugar – lactose – is invariably followed by a second sugar – maltose – soon afterward. Pilpel and his team in the Molecular Genetics Department checked the bacteria’s genetic response to lactose and found that, in addition to the genes that enable it to digest lactose, the gene network for utilizing maltose was partially activated. When they switched the order of the sugars, giving the bacteria maltose first, there was no corresponding activation of lactose genes, implying that bacteria have naturally “learned” to get ready for a serving of maltose after a lactose appetizer.
Another microorganism that experiences consistent changes is wine yeast. As fermentation progresses, sugar and acidity levels change, alcohol levels rise, and the yeast’s environment heats up. Although the system was somewhat more complicated than that of E. coli, the scientists found that when the wine yeast feel the heat, they begin activating genes for dealing with the stresses of the next stage. Further analysis showed that this anticipation and early response is an evolutionary adaptation that increases the organism’s chances of survival.
Ivan Pavlov first demonstrated this type of adaptive anticipation, known as a conditioned response, in dogs in the 1890s. He trained the dogs to salivate in response to a stimulus by repeatedly ringing a bell before giving them food. In the microorganisms, says Pilpel, “evolution over many generations replaces conditioned learning, but the end result is similar.” “In both evolution and learning,” says Mitchell, “the organism adapts its responses to environmental cues, improving its ability to survive.” Romano: “This is not a generalized stress response, but one that is precisely geared to an anticipated event.” To see whether the microorganisms were truly exhibiting a conditioned response, Pilpel and Mitchell devised a further test for the E. coli based on another of Pavlov’s experiments. When Pavlov stopped giving the dogs food after ringing the bell, the conditioned response faded until they eventually ceased salivating at its sound. The scientists did something similar, using bacteria grown by Dr. Erez Dekel, in the lab of Prof. Uri Alon of the Weizmann Institute’s Molecular Cell Biology Department, in an environment containing the first sugar, lactose, but not following it up with maltose. After several months, the bacteria had evolved to stop activating their maltose genes at the taste of lactose, only turning them on when maltose was actually available.
“This showed us that there is a cost to advanced preparation, but that the benefits to the organism outweigh the costs in the right circumstances,” says Pilpel. What are those circumstances? Based on the experimental evidence, the research team created a sort of cost/benefit model to predict the types of situations in which an organism could increase its chances of survival by evolving to anticipate future events. The researchers are already planning a number of new tests for their model, as well as different avenues of experimentation based on the insights they have gained.
Pilpel and his team believe that genetic conditioned response may be a widespread means of evolutionary adaptation that enhances survival in many organisms – one that may also take place in the cells of higher organisms, including humans. These findings could have practical implications, as well. Genetically engineered microorganisms for fermenting plant materials to produce biofuels, for example, might work more efficiently if they gained the genetic ability to prepare themselves for the next step in the process.
Prof. Yitzhak Pilpel’s research is supported by the Ben May Charitable Trust and Madame Huguette Nazez, Paris, France.
The Weizmann Institute of Science in Rehovot, Israel, is one of the world's top-ranking multidisciplinary research institutions. Noted for its wide-ranging exploration of the natural and exact sciences, the Institute is home to 2,600 scientists, students, technicians, and supporting staff. Institute research efforts include the search for new ways of fighting disease and hunger, examining leading questions in mathematics and computer science, probing the physics of matter and the universe, creating novel materials, and developing new strategies for protecting the environment.
Bacteria can anticipate a future event and prepare for it, according to new research at the Weizmann Institute of Science. In a paper that appeared in the June 17, 2009 issue of Nature, Prof. Yitzhak Pilpel, doctoral student Amir Mitchell, and research associate Dr. Orna Dahan of the Institute’s Molecular Genetics Department, together with Prof. Martin Kupiec and Gal Romano of Tel Aviv University, examined microorganisms living in environments that change in predictable ways. Their findings show that these microorganisms’ genetic networks are hard-wired to “foresee” what comes next in the sequence of events and begin responding to the new state of affairs before its onset.
E. coli bacteria, for instance, which normally cruise harmlessly down the digestive tract, encounter a number of different environments on their way. In particular, they find that one type of sugar – lactose – is invariably followed by a second sugar – maltose – soon afterward. Pilpel and his team in the Molecular Genetics Department checked the bacteria’s genetic response to lactose and found that, in addition to the genes that enable it to digest lactose, the gene network for utilizing maltose was partially activated. When they switched the order of the sugars, giving the bacteria maltose first, there was no corresponding activation of lactose genes, implying that bacteria have naturally “learned” to get ready for a serving of maltose after a lactose appetizer.
Another microorganism that experiences consistent changes is wine yeast. As fermentation progresses, sugar and acidity levels change, alcohol levels rise, and the yeast’s environment heats up. Although the system was somewhat more complicated than that of E. coli, the scientists found that when the wine yeast feel the heat, they begin activating genes for dealing with the stresses of the next stage. Further analysis showed that this anticipation and early response is an evolutionary adaptation that increases the organism’s chances of survival.
Ivan Pavlov first demonstrated this type of adaptive anticipation, known as a conditioned response, in dogs in the 1890s. He trained the dogs to salivate in response to a stimulus by repeatedly ringing a bell before giving them food. In the microorganisms, says Pilpel, “evolution over many generations replaces conditioned learning, but the end result is similar.” “In both evolution and learning,” says Mitchell, “the organism adapts its responses to environmental cues, improving its ability to survive.” Romano: “This is not a generalized stress response, but one that is precisely geared to an anticipated event.” To see whether the microorganisms were truly exhibiting a conditioned response, Pilpel and Mitchell devised a further test for the E. coli based on another of Pavlov’s experiments. When Pavlov stopped giving the dogs food after ringing the bell, the conditioned response faded until they eventually ceased salivating at its sound. The scientists did something similar, using bacteria grown by Dr. Erez Dekel, in the lab of Prof. Uri Alon of the Weizmann Institute’s Molecular Cell Biology Department, in an environment containing the first sugar, lactose, but not following it up with maltose. After several months, the bacteria had evolved to stop activating their maltose genes at the taste of lactose, only turning them on when maltose was actually available.
“This showed us that there is a cost to advanced preparation, but that the benefits to the organism outweigh the costs in the right circumstances,” says Pilpel. What are those circumstances? Based on the experimental evidence, the research team created a sort of cost/benefit model to predict the types of situations in which an organism could increase its chances of survival by evolving to anticipate future events. The researchers are already planning a number of new tests for their model, as well as different avenues of experimentation based on the insights they have gained.
Pilpel and his team believe that genetic conditioned response may be a widespread means of evolutionary adaptation that enhances survival in many organisms – one that may also take place in the cells of higher organisms, including humans. These findings could have practical implications, as well. Genetically engineered microorganisms for fermenting plant materials to produce biofuels, for example, might work more efficiently if they gained the genetic ability to prepare themselves for the next step in the process.
Prof. Yitzhak Pilpel’s research is supported by the Ben May Charitable Trust and Madame Huguette Nazez, Paris, France.
The Weizmann Institute of Science in Rehovot, Israel, is one of the world's top-ranking multidisciplinary research institutions. Noted for its wide-ranging exploration of the natural and exact sciences, the Institute is home to 2,600 scientists, students, technicians, and supporting staff. Institute research efforts include the search for new ways of fighting disease and hunger, examining leading questions in mathematics and computer science, probing the physics of matter and the universe, creating novel materials, and developing new strategies for protecting the environment.
Labels:
ADAPTATION,
BACTERIA,
CONDITIONED RESPONSE,
Evolution,
GENETIC ENGINEERING,
PAVLOV,
PILPEL,
WEIZMANN
Key Found to How Tumor Cells Invade the Brain in Childhood Cancer
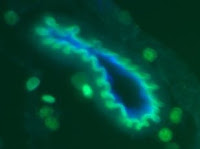 Despite great strides in treating childhood leukemia, a form of the disease called T-cell acute lymphoblastic leukemia (T-ALL) poses special challenges because of the high risk of leukemic cells invading the brain and spinal cord of children who relapse. Now, a new study in the June 18, 2009, issue of the journal Nature by scientists at NYU School of Medicine reveals the molecular agents behind this devastating infiltration of the central nervous system. The finding may lead to new drugs that block these agents and thus lower the risk of relapse.
Despite great strides in treating childhood leukemia, a form of the disease called T-cell acute lymphoblastic leukemia (T-ALL) poses special challenges because of the high risk of leukemic cells invading the brain and spinal cord of children who relapse. Now, a new study in the June 18, 2009, issue of the journal Nature by scientists at NYU School of Medicine reveals the molecular agents behind this devastating infiltration of the central nervous system. The finding may lead to new drugs that block these agents and thus lower the risk of relapse.
T-ALL, a blood-borne cancer in which the bone marrow makes too many lymphocytes, or white blood cells, strikes several hundred children and adolescents in the U.S. annually. While greater than 90% percent go into remission through a combination of chemotherapy and radiation, up to one third of this group end up relapsing. These patients are at particular risk for tumor cells to invade the brain and spinal cord, and to prevent this all patients receive chemotherapy injections into the central nervous system and in some cases cranial irradiation—approaches that cause dangerous side effects, including secondary tumors and potentially permanent cognitive and developmental deficits.
“In general, T-cell acute lymphoblastic leukemia is treatable with chemotherapy and radiation,” said Ioannis Aifantis, PhD, associate professor of pathology and co-director of the Cancer Stem Cell Program at the NYU Cancer Institute, who led the new study. “But you have a very high rate of relapse. And after the relapse, it is not treatable because the cancer occurs in tricky places like the central nervous system,” said Dr. Aifantis, who is also an Early Career Scientist at the Howard Hughes Medical Institute.
“We are very proud of this research and very excited about the potential implications for new therapeutic approaches to prevent or reduce the spread of leukemic cells into the central nervous system,” said Vivian S. Lee MD, PhD, MBA, the vice dean for science, senior vice president and chief scientific officer of NYU Langone Medical Center.
In the new study, Dr. Aifantis and his colleagues found that a key protein receptor embedded on the outer surface of leukemic cells is responsible for infiltrating the brain and spinal cord. “What we have found is that leukemic cells over-express this receptor.” said Dr. Aifantis, “If you knock out this receptor, these cells will not go to the brain under any circumstances.”
Previous research had strongly implicated a famous gene regulator called Notch1 in the progression of T-ALL. The Notch1 gene (a mutated version gives fruit flies notched wings) is an oncogene, or cancer-causing gene, in humans. Certain kinds of mutations in this gene have been found in nearly half of all T-ALL patients, and current estimates suggest that the gene’s regulatory influence might be implicated in nearly 90 percent of all T-ALL cases.
For their new study, Dr. Aifantis and his colleagues first introduced overactive forms of Notch1 into mice. As a result, the mice developed leukemia and the leukemic cells efficiently infiltrated the inner layers of the membrane covering the brain. “What happens is that the leukemic cells get into the cerebrospinal fluidthat protects our brain and spine, where they fill up the space and they can affect brain function, either by secreting chemicals and toxic factors or even by simple pressure,” Dr. Aifantis said.
His team then examined an array of other mouse genes to identify candidates that might fall under the regulatory spell of Notch1 to promote the brain and spinal cord infiltration. The screen revealed a promising gene for a protein named CCR7, which is embedded on the surface of lymphocytes. This chemokine receptor, as it’s known, normally senses and responds to small chemical attractants called chemokines, which act like recruitment signals for lymphocytes to converge on a specific site during the body’s response to infection or injury. In leukemia, however, these lymphocytes proliferate abnormally.

CCR7 was already known as a key player in normal lymphocyte migration and as a binding partner of two chemokines named CCL19 and CCL21. Previous studies had implicated these protein interactions in the metastasis of other tumors such as melanomas and breast cancers. Dr. Aifantis’s team also discovered that the gene for CCR7 was overactive in four of five T-ALL cell lines derived from human patients, bolstering suspicions that it played a central role in the disease. Conversely, a mutation that knocked out Notch1 also led to dramatically reduced CCR7 levels.
To characterize CCR7’s potential role in T-ALL, the researchers used two sets of mice: one in which the receptor was turned on, and a second in which it was turned off. When the team delivered an identical number of human-derived leukemic cells to both sets of mice, those with the CCR7 chemokine receptor turned off lived almost twice as long. Using bioluminescent imaging, the researchers quickly understood why: animals with the active CCR7 receptor had many more tumors. Tellingly, the T-ALL cells had infiltrated the brain and spinal cord of those mice.
Further experiments suggested that when healthy mice received leukemic cells in which the gene for CCR7 had been turned off, the cells could not migrate to the brain even though they reached other body tissues. As a result, the mice survived significantly longer than counterparts with an active copy of the gene. On the other hand, introducing a normal version of the same gene to mice otherwise lacking it was enough to recruit leukemic cells to the brain and spine.
“We wanted to determine whether CCR7 by itself was sufficient for entry into the central nervous system and that’s what this experiment shows,” Dr. Aifantis said. “By changing one specific gene, you now have your function back.”
Finally, the researchers identified the small protein that acted as the “come hither” signal for the CCR7 protein receptors. One candidate, CCL21, was undetectable in leukemic mice. But a second, CCL19, appeared in tiny veins of the brain near the infiltrating tumor cells. When the researchers introduced leukemic cells carrying a gene for CCR7 to mice that naturally lacked the CCL19 chemokine, the mice survived longer, suggesting that their increased life spans might be due to a disrupted interaction of the two proteins. The leukemic cells had no trouble infiltrating other tissue like the lymph nodes, but were completely incapable of infiltrating the brains of CCL19-deficient mice, the researchers report.
“Perhaps there are antibodies or small molecules that can block the interaction between these two proteins or reduce their interactions,” Dr. Aifantis said, “and hopefully that could be used as a type of prophylactic treatment to prevent a relapse in the central nervous system among patients who have already been treated for leukemia.” Such a treatment, he said, could prove a good alternative to the intensive and often poorly tolerated radiation and chemotherapy now used to try to block such a relapse.
The study was led by Dr. Silvia Buonamici, a post-doctoral fellow in the laboratory of Dr. Aifantis in the Department of Pathology and the NYU Cancer Institute, and in the Helen L. and Martin S. Kimmel Stem Cell Center at NYU Langone Medical Center. Other study investigators are; Thomas Trimarchi, Maria Grazia Ruocco, Linsey Reavie, Severine Cathelin, Yevgeniy Lukyanov, Jen-Chieh Tseng, Filiz Sen, Mengling Li, Elizabeth Newcomb, Jiri Zavadil, Daniel Meruelo, Sherif Ibrahim, David Zagzag, and Michael L. Dustin from NYU Langone Medical Center; Brenton G. Mar, Apostolos Klinakis, and Argiris Efstratiadis from Columbia University Medical Center; Eric Gehrie and Jonathan S. Bromberg from Mount Sinai School of Medicine; and Martin Lipp from the Max Delbrück Center for Molecular Medicine in Berlin.
The study was supported by grants from the National Institutes of Health, the American Cancer Society, the Dana Foundation, The Chemotherapy Foundation, the Alex’s Lemonade Stand Foundation, the Lauri Strauss Leukemia foundation, the G&P Foundation, an NYU School of Medicine Molecular Oncology and Immunology training grant, the American Society of Hematology, the Juvenile Diabetes Research Foundation, the National Cancer Institute, a gift from the Berrie Foundation, and a fellowship from the Jane Coffin Childs Memorial Fund for Medical Research.
About NYU Langone Medical Center
Located in New York City, NYU Langone Medical Center is one of the nation's premier centers of excellence in health care, biomedical research, and medical education. For over 168 years, NYU physicians and researchers have made countless contributions to the practice and science of health care. Today the Medical Center consists of NYU School of Medicine, including the Smilow Research Center, the Skirball Institute of Biomolecular Medicine, and the Sackler Institute of Graduate Biomedical Sciences; the three hospitals of NYU Hospitals Center, Tisch Hospital, a 705-bed acute-care general hospital, Rusk Institute of Rehabilitation Medicine, the first and largest facility of its kind, and NYU Hospital for Joint Diseases, a leader in musculoskeletal care; and such major programs as the NYU Cancer Institute, the NYU Child Study Center, and the Hassenfeld Children's Center for Cancer and Blood Disorders.
About NYU Cancer Institute
The NYU Cancer Institute is an NCI-designated cancer center. Its mission is to discover the origins of human cancer and to use that knowledge to eradicate the personal and societal burden of cancer in our community, the nation and the world. The center and its multidisciplinary team of experts provide access to the latest treatment options and clinical trials along with a variety of programs in cancer prevention, screening, diagnostics, genetic counseling and supportive services. For additional information, please visit: www.nyuci.org.
 Despite great strides in treating childhood leukemia, a form of the disease called T-cell acute lymphoblastic leukemia (T-ALL) poses special challenges because of the high risk of leukemic cells invading the brain and spinal cord of children who relapse. Now, a new study in the June 18, 2009, issue of the journal Nature by scientists at NYU School of Medicine reveals the molecular agents behind this devastating infiltration of the central nervous system. The finding may lead to new drugs that block these agents and thus lower the risk of relapse.
Despite great strides in treating childhood leukemia, a form of the disease called T-cell acute lymphoblastic leukemia (T-ALL) poses special challenges because of the high risk of leukemic cells invading the brain and spinal cord of children who relapse. Now, a new study in the June 18, 2009, issue of the journal Nature by scientists at NYU School of Medicine reveals the molecular agents behind this devastating infiltration of the central nervous system. The finding may lead to new drugs that block these agents and thus lower the risk of relapse. T-ALL, a blood-borne cancer in which the bone marrow makes too many lymphocytes, or white blood cells, strikes several hundred children and adolescents in the U.S. annually. While greater than 90% percent go into remission through a combination of chemotherapy and radiation, up to one third of this group end up relapsing. These patients are at particular risk for tumor cells to invade the brain and spinal cord, and to prevent this all patients receive chemotherapy injections into the central nervous system and in some cases cranial irradiation—approaches that cause dangerous side effects, including secondary tumors and potentially permanent cognitive and developmental deficits.
“In general, T-cell acute lymphoblastic leukemia is treatable with chemotherapy and radiation,” said Ioannis Aifantis, PhD, associate professor of pathology and co-director of the Cancer Stem Cell Program at the NYU Cancer Institute, who led the new study. “But you have a very high rate of relapse. And after the relapse, it is not treatable because the cancer occurs in tricky places like the central nervous system,” said Dr. Aifantis, who is also an Early Career Scientist at the Howard Hughes Medical Institute.
“We are very proud of this research and very excited about the potential implications for new therapeutic approaches to prevent or reduce the spread of leukemic cells into the central nervous system,” said Vivian S. Lee MD, PhD, MBA, the vice dean for science, senior vice president and chief scientific officer of NYU Langone Medical Center.
In the new study, Dr. Aifantis and his colleagues found that a key protein receptor embedded on the outer surface of leukemic cells is responsible for infiltrating the brain and spinal cord. “What we have found is that leukemic cells over-express this receptor.” said Dr. Aifantis, “If you knock out this receptor, these cells will not go to the brain under any circumstances.”
Previous research had strongly implicated a famous gene regulator called Notch1 in the progression of T-ALL. The Notch1 gene (a mutated version gives fruit flies notched wings) is an oncogene, or cancer-causing gene, in humans. Certain kinds of mutations in this gene have been found in nearly half of all T-ALL patients, and current estimates suggest that the gene’s regulatory influence might be implicated in nearly 90 percent of all T-ALL cases.
For their new study, Dr. Aifantis and his colleagues first introduced overactive forms of Notch1 into mice. As a result, the mice developed leukemia and the leukemic cells efficiently infiltrated the inner layers of the membrane covering the brain. “What happens is that the leukemic cells get into the cerebrospinal fluidthat protects our brain and spine, where they fill up the space and they can affect brain function, either by secreting chemicals and toxic factors or even by simple pressure,” Dr. Aifantis said.
His team then examined an array of other mouse genes to identify candidates that might fall under the regulatory spell of Notch1 to promote the brain and spinal cord infiltration. The screen revealed a promising gene for a protein named CCR7, which is embedded on the surface of lymphocytes. This chemokine receptor, as it’s known, normally senses and responds to small chemical attractants called chemokines, which act like recruitment signals for lymphocytes to converge on a specific site during the body’s response to infection or injury. In leukemia, however, these lymphocytes proliferate abnormally.

CCR7 was already known as a key player in normal lymphocyte migration and as a binding partner of two chemokines named CCL19 and CCL21. Previous studies had implicated these protein interactions in the metastasis of other tumors such as melanomas and breast cancers. Dr. Aifantis’s team also discovered that the gene for CCR7 was overactive in four of five T-ALL cell lines derived from human patients, bolstering suspicions that it played a central role in the disease. Conversely, a mutation that knocked out Notch1 also led to dramatically reduced CCR7 levels.
To characterize CCR7’s potential role in T-ALL, the researchers used two sets of mice: one in which the receptor was turned on, and a second in which it was turned off. When the team delivered an identical number of human-derived leukemic cells to both sets of mice, those with the CCR7 chemokine receptor turned off lived almost twice as long. Using bioluminescent imaging, the researchers quickly understood why: animals with the active CCR7 receptor had many more tumors. Tellingly, the T-ALL cells had infiltrated the brain and spinal cord of those mice.
Further experiments suggested that when healthy mice received leukemic cells in which the gene for CCR7 had been turned off, the cells could not migrate to the brain even though they reached other body tissues. As a result, the mice survived significantly longer than counterparts with an active copy of the gene. On the other hand, introducing a normal version of the same gene to mice otherwise lacking it was enough to recruit leukemic cells to the brain and spine.
“We wanted to determine whether CCR7 by itself was sufficient for entry into the central nervous system and that’s what this experiment shows,” Dr. Aifantis said. “By changing one specific gene, you now have your function back.”
Finally, the researchers identified the small protein that acted as the “come hither” signal for the CCR7 protein receptors. One candidate, CCL21, was undetectable in leukemic mice. But a second, CCL19, appeared in tiny veins of the brain near the infiltrating tumor cells. When the researchers introduced leukemic cells carrying a gene for CCR7 to mice that naturally lacked the CCL19 chemokine, the mice survived longer, suggesting that their increased life spans might be due to a disrupted interaction of the two proteins. The leukemic cells had no trouble infiltrating other tissue like the lymph nodes, but were completely incapable of infiltrating the brains of CCL19-deficient mice, the researchers report.
“Perhaps there are antibodies or small molecules that can block the interaction between these two proteins or reduce their interactions,” Dr. Aifantis said, “and hopefully that could be used as a type of prophylactic treatment to prevent a relapse in the central nervous system among patients who have already been treated for leukemia.” Such a treatment, he said, could prove a good alternative to the intensive and often poorly tolerated radiation and chemotherapy now used to try to block such a relapse.
The study was led by Dr. Silvia Buonamici, a post-doctoral fellow in the laboratory of Dr. Aifantis in the Department of Pathology and the NYU Cancer Institute, and in the Helen L. and Martin S. Kimmel Stem Cell Center at NYU Langone Medical Center. Other study investigators are; Thomas Trimarchi, Maria Grazia Ruocco, Linsey Reavie, Severine Cathelin, Yevgeniy Lukyanov, Jen-Chieh Tseng, Filiz Sen, Mengling Li, Elizabeth Newcomb, Jiri Zavadil, Daniel Meruelo, Sherif Ibrahim, David Zagzag, and Michael L. Dustin from NYU Langone Medical Center; Brenton G. Mar, Apostolos Klinakis, and Argiris Efstratiadis from Columbia University Medical Center; Eric Gehrie and Jonathan S. Bromberg from Mount Sinai School of Medicine; and Martin Lipp from the Max Delbrück Center for Molecular Medicine in Berlin.
The study was supported by grants from the National Institutes of Health, the American Cancer Society, the Dana Foundation, The Chemotherapy Foundation, the Alex’s Lemonade Stand Foundation, the Lauri Strauss Leukemia foundation, the G&P Foundation, an NYU School of Medicine Molecular Oncology and Immunology training grant, the American Society of Hematology, the Juvenile Diabetes Research Foundation, the National Cancer Institute, a gift from the Berrie Foundation, and a fellowship from the Jane Coffin Childs Memorial Fund for Medical Research.
About NYU Langone Medical Center
Located in New York City, NYU Langone Medical Center is one of the nation's premier centers of excellence in health care, biomedical research, and medical education. For over 168 years, NYU physicians and researchers have made countless contributions to the practice and science of health care. Today the Medical Center consists of NYU School of Medicine, including the Smilow Research Center, the Skirball Institute of Biomolecular Medicine, and the Sackler Institute of Graduate Biomedical Sciences; the three hospitals of NYU Hospitals Center, Tisch Hospital, a 705-bed acute-care general hospital, Rusk Institute of Rehabilitation Medicine, the first and largest facility of its kind, and NYU Hospital for Joint Diseases, a leader in musculoskeletal care; and such major programs as the NYU Cancer Institute, the NYU Child Study Center, and the Hassenfeld Children's Center for Cancer and Blood Disorders.
About NYU Cancer Institute
The NYU Cancer Institute is an NCI-designated cancer center. Its mission is to discover the origins of human cancer and to use that knowledge to eradicate the personal and societal burden of cancer in our community, the nation and the world. The center and its multidisciplinary team of experts provide access to the latest treatment options and clinical trials along with a variety of programs in cancer prevention, screening, diagnostics, genetic counseling and supportive services. For additional information, please visit: www.nyuci.org.
Tuesday, June 09, 2009
Confusion About Sugars
Three top researchers corrected inaccuracies and misunderstandings concerning high fructose corn syrup's impact on the American diet. They also examined how the United States Department of Agriculture (USDA) considers this sweetener in light of the upcoming 2010 Dietary Guidelines for Americans in a session, High Fructose Corn Syrup: Sorting Myth from Reality, at the Institute of Food Technologists (IFT) Annual Meeting in Anaheim, California.
"Contrary to its name, high fructose corn syrup is essentially a corn sugar," stated sweetener expert John S. White, Ph.D., president of White Technical Research. "Recent marketing claims that sugar is healthier than high fructose corn syrup are misleading to consumers."
"By every parameter yet measured in human beings, high fructose corn syrup and sugar are identical. This is not surprising since high fructose corn syrup and sugar are metabolized the same by the body, have the same level of sweetness and the same number of calories per gram," noted James M. Rippe, M.D., cardiologist and biomedical sciences professor at the University of Central Florida.
"This is a marketing issue, not a metabolic issue," stated David Klurfeld, Ph.D., national program leader for human nutrition in USDA's Agricultural Research Service and editor of the June 2009 Journal of Nutrition supplement, "The State of the Science on Dietary Sweeteners Containing Fructose," in response to recent reformulations by manufacturers of products that once contained high fructose corn syrup. "The real issue is not high fructose corn syrup. It's that we've forgotten what a real serving size is. We have to eat less of everything," he noted.
Increased Caloric Intake, Not a Single Sweetener, the Likely Cause of Obesity
Fructose-containing sweeteners -- such as sugar, invert sugar, honey, fruit juice concentrates, and high fructose corn syrup -- are essentially interchangeable in composition, calories, and metabolism. Replacing high fructose corn syrup in foods with other fructose-containing sweeteners will provide neither improved nutrition nor a meaningful solution to the obesity crisis, according to Dr. White. "In light of similarities in composition, sweetness, energy content, processing, and metabolism, claims that such sweetener substitutions bring nutritional benefit to children and their families appear disingenuous and misguided," White says.
Growing Body of Evidence
The American Medical Association helped put to rest a common misunderstanding about high fructose corn syrup and obesity, stating that "high fructose syrup does not appear to contribute to obesity more than other caloric sweeteners." Even former critics of high fructose corn syrup dispelled myths and distanced themselves from earlier speculation about the sweetener's link to obesity in a comprehensive scientific review published in the December 2008 American Journal of Clinical Nutrition.
Three top researchers corrected inaccuracies and misunderstandings concerning high fructose corn syrup's impact on the American diet. They also examined how the United States Department of Agriculture (USDA) considers this sweetener in light of the upcoming 2010 Dietary Guidelines for Americans in a session, High Fructose Corn Syrup: Sorting Myth from Reality, at the Institute of Food Technologists (IFT) Annual Meeting in Anaheim, California.
"Contrary to its name, high fructose corn syrup is essentially a corn sugar," stated sweetener expert John S. White, Ph.D., president of White Technical Research. "Recent marketing claims that sugar is healthier than high fructose corn syrup are misleading to consumers."
"By every parameter yet measured in human beings, high fructose corn syrup and sugar are identical. This is not surprising since high fructose corn syrup and sugar are metabolized the same by the body, have the same level of sweetness and the same number of calories per gram," noted James M. Rippe, M.D., cardiologist and biomedical sciences professor at the University of Central Florida.
"This is a marketing issue, not a metabolic issue," stated David Klurfeld, Ph.D., national program leader for human nutrition in USDA's Agricultural Research Service and editor of the June 2009 Journal of Nutrition supplement, "The State of the Science on Dietary Sweeteners Containing Fructose," in response to recent reformulations by manufacturers of products that once contained high fructose corn syrup. "The real issue is not high fructose corn syrup. It's that we've forgotten what a real serving size is. We have to eat less of everything," he noted.
Increased Caloric Intake, Not a Single Sweetener, the Likely Cause of Obesity
Fructose-containing sweeteners -- such as sugar, invert sugar, honey, fruit juice concentrates, and high fructose corn syrup -- are essentially interchangeable in composition, calories, and metabolism. Replacing high fructose corn syrup in foods with other fructose-containing sweeteners will provide neither improved nutrition nor a meaningful solution to the obesity crisis, according to Dr. White. "In light of similarities in composition, sweetness, energy content, processing, and metabolism, claims that such sweetener substitutions bring nutritional benefit to children and their families appear disingenuous and misguided," White says.
Growing Body of Evidence
The American Medical Association helped put to rest a common misunderstanding about high fructose corn syrup and obesity, stating that "high fructose syrup does not appear to contribute to obesity more than other caloric sweeteners." Even former critics of high fructose corn syrup dispelled myths and distanced themselves from earlier speculation about the sweetener's link to obesity in a comprehensive scientific review published in the December 2008 American Journal of Clinical Nutrition.
Saturday, May 02, 2009
Researchers Construct Carbon Nanotube Device That Can Detect Colors of the Rainbow
Researchers at Sandia National Laboratories have created the first carbon nanotube device that can detect the entire visible spectrum of light, a feat that could soon allow scientists to probe single molecule transformations, study how those molecules respond to light, observe how the molecules change shapes, and understand other fundamental interactions between molecules and nanotubes.
Carbon nanotubes are long thin cylinders composed entirely of carbon atoms. While their diameters are in the nanometer range (1-10), they can be very long, up to centimeters in length.
The carbon-carbon bond is very strong, making carbon nanotubes very robust and resistant to any kind of deformation. To construct a nanoscale color detector, Sandia researchers took inspiration from the human eye, and in a sense, improved on the model.
When light strikes the retina, it initiates a cascade of chemical and electrical impulses that ultimately trigger nerve impulses. In the nanoscale color detector, light strikes a chromophore and causes a conformational change in the molecule, which in turn causes a threshold shift on a transistor made from a single-walled carbon nanotube.
“In our eyes the neuron is in front of the retinal molecule, so the light has to transmit through the neuron to hit the molecule,” says Sandia researcher Xinjian Zhou. “We placed the nanotube transistor behind the molecule—a more efficient design.”
Zhou and his Sandia colleagues François Léonard, Andy Vance, Karen Krafcik, Tom Zifer, and Bryan Wong created the device. The team recently published a paper, “Color Detection Using Chromophore-Nanotube Hybrid Devices,” in the journal Nano Letters.
The idea of carbon nanotubes being light sensitive has been around for a long time, but earlier efforts using an individual nanotube were only able to detect light in narrow wavelength ranges at laser intensities. The Sandia team found that their nanodetector was orders of magnitude more sensitive, down to about 40 W/m2—about 3 percent of the density of sunshine reaching the ground. “Because the dye is so close to the nanotube, a little change turns into a big signal on the device,” says Zhou.
The research is in its second year of internal Sandia funding and is based on Léonard’s collaboration with the University of Wisconsin to explain the theoretical mechanism of carbon nanotube light detection. Léonard literally wrote the book on carbon nanotubes—The Physics of Carbon Nanotubes, published September 2008.
Léonard says the project draws upon Sandia’s expertise in both materials physics and materials chemistry. He and Wong laid the groundwork with their theoretical research, with Wong completing the first-principles calculations that supported the hypothesis of how the chromophores were arranged on the nanotubes and how the chromophore isomerizations affected electronic properties of the devices.
To construct the device, Zhou and Krafcik first had to create a tiny transistor made from a single carbon nanotube. They deposited carbon nanotubes on a silicon wafer and then used photolithography to define electrical patterns to make contacts.
The final piece came from Vance and Zifer, who synthesized molecules to create three types of chromophores that respond to either the red, green, or orange bands of the visible spectrum. Zhou immersed the wafer in the dye solution and waited a few minutes while the chromophores attached themselves to the nanotubes.
The team reached their goal of detecting visible light faster than they expected—they thought the entire first year of the project would be spent testing UV light. Now, they are looking to increase the efficiency by creating a device with multiple nanotubes.
“Detection is now limited to about 3 percent of sunlight, which isn’t bad compared with a commercially available digital camera,” says Zhou. “I hope to add some antennas to increase light absorption.”
A device made with multiple carbon nanotubes would be easier to construct and the resulting larger area would be more sensitive to light. A larger size is also more practical for applications.
Now, they are setting their sites on detecting infrared light. “We think this principle can be applied to infrared light and there is a lot of interest in infrared detection,” says Vance. “So we’re in the process of looking for dyes that work in infrared.”
This research eventually could be used for a number of exciting applications, such as an optical detector with nanometer scale resolution, ultra-tiny digital cameras, solar cells with more light absorption capability, or even genome sequencing. The near-term purpose, however, is basic science.
“A large part of why we are doing this is not to invent a photo detector, but to understand the processes involved in controlling carbon nanotube devices,” says Léonard.
The next step in the project is to create a nanometer-scale photovoltaic device. Such a device on a larger scale could be used as an unpowered photo detector or for solar energy. “Instead of monitoring current changes, we’d actually generate current,” says Vance. “We have an idea of how to do it, but it will be a more challenging fabrication process.”
--------------------------------------------------------------------------------
Sandia is a multiprogram laboratory operated by Sandia Corporation, a Lockheed Martin company, for the U.S. Department of Energy’s National Nuclear Security Administration. With main facilities in Albuquerque, N.M., and Livermore, Calif.Researchers at Sandia National Laboratories have created the first carbon nanotube device that can detect the entire visible spectrum of light, a feat that could soon allow scientists to probe single molecule transformations, study how those molecules respond to light, observe how the molecules change shapes, and understand other fundamental interactions between molecules and nanotubes.
Carbon nanotubes are long thin cylinders composed entirely of carbon atoms. While their diameters are in the nanometer range (1-10), they can be very long, up to centimeters in length.
The carbon-carbon bond is very strong, making carbon nanotubes very robust and resistant to any kind of deformation. To construct a nanoscale color detector, Sandia researchers took inspiration from the human eye, and in a sense, improved on the model.
When light strikes the retina, it initiates a cascade of chemical and electrical impulses that ultimately trigger nerve impulses. In the nanoscale color detector, light strikes a chromophore and causes a conformational change in the molecule, which in turn causes a threshold shift on a transistor made from a single-walled carbon nanotube.
“In our eyes the neuron is in front of the retinal molecule, so the light has to transmit through the neuron to hit the molecule,” says Sandia researcher Xinjian Zhou. “We placed the nanotube transistor behind the molecule—a more efficient design.”
Zhou and his Sandia colleagues François Léonard, Andy Vance, Karen Krafcik, Tom Zifer, and Bryan Wong created the device. The team recently published a paper, “Color Detection Using Chromophore-Nanotube Hybrid Devices,” in the journal Nano Letters.
The idea of carbon nanotubes being light sensitive has been around for a long time, but earlier efforts using an individual nanotube were only able to detect light in narrow wavelength ranges at laser intensities. The Sandia team found that their nanodetector was orders of magnitude more sensitive, down to about 40 W/m2—about 3 percent of the density of sunshine reaching the ground. “Because the dye is so close to the nanotube, a little change turns into a big signal on the device,” says Zhou.
The research is in its second year of internal Sandia funding and is based on Léonard’s collaboration with the University of Wisconsin to explain the theoretical mechanism of carbon nanotube light detection. Léonard literally wrote the book on carbon nanotubes—The Physics of Carbon Nanotubes, published September 2008.
Léonard says the project draws upon Sandia’s expertise in both materials physics and materials chemistry. He and Wong laid the groundwork with their theoretical research, with Wong completing the first-principles calculations that supported the hypothesis of how the chromophores were arranged on the nanotubes and how the chromophore isomerizations affected electronic properties of the devices.
To construct the device, Zhou and Krafcik first had to create a tiny transistor made from a single carbon nanotube. They deposited carbon nanotubes on a silicon wafer and then used photolithography to define electrical patterns to make contacts.
The final piece came from Vance and Zifer, who synthesized molecules to create three types of chromophores that respond to either the red, green, or orange bands of the visible spectrum. Zhou immersed the wafer in the dye solution and waited a few minutes while the chromophores attached themselves to the nanotubes.
The team reached their goal of detecting visible light faster than they expected—they thought the entire first year of the project would be spent testing UV light. Now, they are looking to increase the efficiency by creating a device with multiple nanotubes.
“Detection is now limited to about 3 percent of sunlight, which isn’t bad compared with a commercially available digital camera,” says Zhou. “I hope to add some antennas to increase light absorption.”
A device made with multiple carbon nanotubes would be easier to construct and the resulting larger area would be more sensitive to light. A larger size is also more practical for applications.
Now, they are setting their sites on detecting infrared light. “We think this principle can be applied to infrared light and there is a lot of interest in infrared detection,” says Vance. “So we’re in the process of looking for dyes that work in infrared.”
This research eventually could be used for a number of exciting applications, such as an optical detector with nanometer scale resolution, ultra-tiny digital cameras, solar cells with more light absorption capability, or even genome sequencing. The near-term purpose, however, is basic science.
“A large part of why we are doing this is not to invent a photo detector, but to understand the processes involved in controlling carbon nanotube devices,” says Léonard.
The next step in the project is to create a nanometer-scale photovoltaic device. Such a device on a larger scale could be used as an unpowered photo detector or for solar energy. “Instead of monitoring current changes, we’d actually generate current,” says Vance. “We have an idea of how to do it, but it will be a more challenging fabrication process.”
--------------------------------------------------------------------------------
Sandia is a multiprogram laboratory operated by Sandia Corporation, a Lockheed Martin company, for the U.S. Department of Energy’s National Nuclear Security Administration. With main facilities in Albuquerque, N.M., and Livermore, Calif.
Sandia has major R&D responsibilities in national security, energy and environmental technologies, and economic competitiveness.
Researchers at Sandia National Laboratories have created the first carbon nanotube device that can detect the entire visible spectrum of light, a feat that could soon allow scientists to probe single molecule transformations, study how those molecules respond to light, observe how the molecules change shapes, and understand other fundamental interactions between molecules and nanotubes.
Carbon nanotubes are long thin cylinders composed entirely of carbon atoms. While their diameters are in the nanometer range (1-10), they can be very long, up to centimeters in length.
The carbon-carbon bond is very strong, making carbon nanotubes very robust and resistant to any kind of deformation. To construct a nanoscale color detector, Sandia researchers took inspiration from the human eye, and in a sense, improved on the model.
When light strikes the retina, it initiates a cascade of chemical and electrical impulses that ultimately trigger nerve impulses. In the nanoscale color detector, light strikes a chromophore and causes a conformational change in the molecule, which in turn causes a threshold shift on a transistor made from a single-walled carbon nanotube.
“In our eyes the neuron is in front of the retinal molecule, so the light has to transmit through the neuron to hit the molecule,” says Sandia researcher Xinjian Zhou. “We placed the nanotube transistor behind the molecule—a more efficient design.”
Zhou and his Sandia colleagues François Léonard, Andy Vance, Karen Krafcik, Tom Zifer, and Bryan Wong created the device. The team recently published a paper, “Color Detection Using Chromophore-Nanotube Hybrid Devices,” in the journal Nano Letters.
The idea of carbon nanotubes being light sensitive has been around for a long time, but earlier efforts using an individual nanotube were only able to detect light in narrow wavelength ranges at laser intensities. The Sandia team found that their nanodetector was orders of magnitude more sensitive, down to about 40 W/m2—about 3 percent of the density of sunshine reaching the ground. “Because the dye is so close to the nanotube, a little change turns into a big signal on the device,” says Zhou.
The research is in its second year of internal Sandia funding and is based on Léonard’s collaboration with the University of Wisconsin to explain the theoretical mechanism of carbon nanotube light detection. Léonard literally wrote the book on carbon nanotubes—The Physics of Carbon Nanotubes, published September 2008.
Léonard says the project draws upon Sandia’s expertise in both materials physics and materials chemistry. He and Wong laid the groundwork with their theoretical research, with Wong completing the first-principles calculations that supported the hypothesis of how the chromophores were arranged on the nanotubes and how the chromophore isomerizations affected electronic properties of the devices.
To construct the device, Zhou and Krafcik first had to create a tiny transistor made from a single carbon nanotube. They deposited carbon nanotubes on a silicon wafer and then used photolithography to define electrical patterns to make contacts.
The final piece came from Vance and Zifer, who synthesized molecules to create three types of chromophores that respond to either the red, green, or orange bands of the visible spectrum. Zhou immersed the wafer in the dye solution and waited a few minutes while the chromophores attached themselves to the nanotubes.
The team reached their goal of detecting visible light faster than they expected—they thought the entire first year of the project would be spent testing UV light. Now, they are looking to increase the efficiency by creating a device with multiple nanotubes.
“Detection is now limited to about 3 percent of sunlight, which isn’t bad compared with a commercially available digital camera,” says Zhou. “I hope to add some antennas to increase light absorption.”
A device made with multiple carbon nanotubes would be easier to construct and the resulting larger area would be more sensitive to light. A larger size is also more practical for applications.
Now, they are setting their sites on detecting infrared light. “We think this principle can be applied to infrared light and there is a lot of interest in infrared detection,” says Vance. “So we’re in the process of looking for dyes that work in infrared.”
This research eventually could be used for a number of exciting applications, such as an optical detector with nanometer scale resolution, ultra-tiny digital cameras, solar cells with more light absorption capability, or even genome sequencing. The near-term purpose, however, is basic science.
“A large part of why we are doing this is not to invent a photo detector, but to understand the processes involved in controlling carbon nanotube devices,” says Léonard.
The next step in the project is to create a nanometer-scale photovoltaic device. Such a device on a larger scale could be used as an unpowered photo detector or for solar energy. “Instead of monitoring current changes, we’d actually generate current,” says Vance. “We have an idea of how to do it, but it will be a more challenging fabrication process.”
--------------------------------------------------------------------------------
Sandia is a multiprogram laboratory operated by Sandia Corporation, a Lockheed Martin company, for the U.S. Department of Energy’s National Nuclear Security Administration. With main facilities in Albuquerque, N.M., and Livermore, Calif.Researchers at Sandia National Laboratories have created the first carbon nanotube device that can detect the entire visible spectrum of light, a feat that could soon allow scientists to probe single molecule transformations, study how those molecules respond to light, observe how the molecules change shapes, and understand other fundamental interactions between molecules and nanotubes.
Carbon nanotubes are long thin cylinders composed entirely of carbon atoms. While their diameters are in the nanometer range (1-10), they can be very long, up to centimeters in length.
The carbon-carbon bond is very strong, making carbon nanotubes very robust and resistant to any kind of deformation. To construct a nanoscale color detector, Sandia researchers took inspiration from the human eye, and in a sense, improved on the model.
When light strikes the retina, it initiates a cascade of chemical and electrical impulses that ultimately trigger nerve impulses. In the nanoscale color detector, light strikes a chromophore and causes a conformational change in the molecule, which in turn causes a threshold shift on a transistor made from a single-walled carbon nanotube.
“In our eyes the neuron is in front of the retinal molecule, so the light has to transmit through the neuron to hit the molecule,” says Sandia researcher Xinjian Zhou. “We placed the nanotube transistor behind the molecule—a more efficient design.”
Zhou and his Sandia colleagues François Léonard, Andy Vance, Karen Krafcik, Tom Zifer, and Bryan Wong created the device. The team recently published a paper, “Color Detection Using Chromophore-Nanotube Hybrid Devices,” in the journal Nano Letters.
The idea of carbon nanotubes being light sensitive has been around for a long time, but earlier efforts using an individual nanotube were only able to detect light in narrow wavelength ranges at laser intensities. The Sandia team found that their nanodetector was orders of magnitude more sensitive, down to about 40 W/m2—about 3 percent of the density of sunshine reaching the ground. “Because the dye is so close to the nanotube, a little change turns into a big signal on the device,” says Zhou.
The research is in its second year of internal Sandia funding and is based on Léonard’s collaboration with the University of Wisconsin to explain the theoretical mechanism of carbon nanotube light detection. Léonard literally wrote the book on carbon nanotubes—The Physics of Carbon Nanotubes, published September 2008.
Léonard says the project draws upon Sandia’s expertise in both materials physics and materials chemistry. He and Wong laid the groundwork with their theoretical research, with Wong completing the first-principles calculations that supported the hypothesis of how the chromophores were arranged on the nanotubes and how the chromophore isomerizations affected electronic properties of the devices.
To construct the device, Zhou and Krafcik first had to create a tiny transistor made from a single carbon nanotube. They deposited carbon nanotubes on a silicon wafer and then used photolithography to define electrical patterns to make contacts.
The final piece came from Vance and Zifer, who synthesized molecules to create three types of chromophores that respond to either the red, green, or orange bands of the visible spectrum. Zhou immersed the wafer in the dye solution and waited a few minutes while the chromophores attached themselves to the nanotubes.
The team reached their goal of detecting visible light faster than they expected—they thought the entire first year of the project would be spent testing UV light. Now, they are looking to increase the efficiency by creating a device with multiple nanotubes.
“Detection is now limited to about 3 percent of sunlight, which isn’t bad compared with a commercially available digital camera,” says Zhou. “I hope to add some antennas to increase light absorption.”
A device made with multiple carbon nanotubes would be easier to construct and the resulting larger area would be more sensitive to light. A larger size is also more practical for applications.
Now, they are setting their sites on detecting infrared light. “We think this principle can be applied to infrared light and there is a lot of interest in infrared detection,” says Vance. “So we’re in the process of looking for dyes that work in infrared.”
This research eventually could be used for a number of exciting applications, such as an optical detector with nanometer scale resolution, ultra-tiny digital cameras, solar cells with more light absorption capability, or even genome sequencing. The near-term purpose, however, is basic science.
“A large part of why we are doing this is not to invent a photo detector, but to understand the processes involved in controlling carbon nanotube devices,” says Léonard.
The next step in the project is to create a nanometer-scale photovoltaic device. Such a device on a larger scale could be used as an unpowered photo detector or for solar energy. “Instead of monitoring current changes, we’d actually generate current,” says Vance. “We have an idea of how to do it, but it will be a more challenging fabrication process.”
--------------------------------------------------------------------------------
Sandia is a multiprogram laboratory operated by Sandia Corporation, a Lockheed Martin company, for the U.S. Department of Energy’s National Nuclear Security Administration. With main facilities in Albuquerque, N.M., and Livermore, Calif.
Sandia has major R&D responsibilities in national security, energy and environmental technologies, and economic competitiveness.
Labels:
COLOR,
DETECTION,
NANO,
Nanotechnology,
SANDIA LABS
Saturday, April 25, 2009
Vitamin D 'is a hormone'
A new study has suggested that vitamin D isn't really a vitamin at all -- it's actually a hormone made inside the body without any help from the sun.
An international team has carried out the study and concluded that the increase of vitamin D in our modern diets is based on a common belief which is actually a misconception with potential consequences.
ÒWhat we have confirmed with our recent research is that vitamin D is a hormone that is made by the body itself. Our bodies hormonal control system was being overwhelmed by the amount of external vitamin D,Ó lead researcher Prof Trevor G Marshall at Murdoch University in Australia said.
The researchers go on to explode another long held belief about this secosteriod previously known as vitamin D. ÒYou don't have to ingest any vitamin D in order to be perfectly healthy,Ó Prof Marshall said.
So no more need for expensive supplements, no more basking in the sun to put us in a better mood? And what about the thinking that suggests vitamin D is vital in production of serotonin, an essential element linked to helping maintaining normal brain chemical function? ÓWhat we've shown is that all forms of vitamin D from outside the body are counterproductive to body's own ability to regulate its own internal production,Ó he said.
This conclusion doesn't mean a dramatic change of lifestyle where we must all suddenly shun the sun but the researchers do acknowledge that people have only been at risk of vitamin D overexposure from about the same time as when bikinis made an appearance.
ÒHistorically the amount of sunshine which people have typically been getting was adequate, certainly up until the mid twentieth century when we started to do silly things like sunbathing and wearing bikinis, and before that time people were already sourcing enough vitamin D from everyday foods like fish, mushrooms and eggs,Ó Prof Marshall said.
A new study has suggested that vitamin D isn't really a vitamin at all -- it's actually a hormone made inside the body without any help from the sun.
An international team has carried out the study and concluded that the increase of vitamin D in our modern diets is based on a common belief which is actually a misconception with potential consequences.
ÒWhat we have confirmed with our recent research is that vitamin D is a hormone that is made by the body itself. Our bodies hormonal control system was being overwhelmed by the amount of external vitamin D,Ó lead researcher Prof Trevor G Marshall at Murdoch University in Australia said.
The researchers go on to explode another long held belief about this secosteriod previously known as vitamin D. ÒYou don't have to ingest any vitamin D in order to be perfectly healthy,Ó Prof Marshall said.
So no more need for expensive supplements, no more basking in the sun to put us in a better mood? And what about the thinking that suggests vitamin D is vital in production of serotonin, an essential element linked to helping maintaining normal brain chemical function? ÓWhat we've shown is that all forms of vitamin D from outside the body are counterproductive to body's own ability to regulate its own internal production,Ó he said.
This conclusion doesn't mean a dramatic change of lifestyle where we must all suddenly shun the sun but the researchers do acknowledge that people have only been at risk of vitamin D overexposure from about the same time as when bikinis made an appearance.
ÒHistorically the amount of sunshine which people have typically been getting was adequate, certainly up until the mid twentieth century when we started to do silly things like sunbathing and wearing bikinis, and before that time people were already sourcing enough vitamin D from everyday foods like fish, mushrooms and eggs,Ó Prof Marshall said.
The World’s First, High Performance & Environmentally Benign Power Generation Unit
If the device is applied to 30% of the cars in Japan, we can expect a 960,000kl oil substitution effect which is 1.5 times the assumed effect from solar energy generation of the year 2010.
A research team led by associate professor Tsutomu Iida (Tokyo University of Science - Faculty of Industrial Science and Technology – Materials Science and Technology) has developed a power generation unit driven by wasted heat composed of Magnesium silicide (Mg2Si), a filtered by-product of Si-LSI and solar cell cast wafer production.
This device, being the first in the world, has been successful in bulk-quantity synthesis, at the same time increasing the thermoelectric conversion rate. Approximately 2,500W/m² (per unit) of power generation and 3,000 hrs of continuous operation has been made possible, sufficiently fulfilling the criteria for commercial use.
By applying this device in industrial shaft furnaces and/or car engines, we can expect drastic reduction in fuel consumption and prevention of global warming. Implementation of this device has already been determined partially as an experiment for practical use and there are high expectations of application of this device in industrial furnaces nation-wide.
Mg2Si Power Generation Unit Driven by Waste Heat - Capability & Effect; This device has an effective temperature range from 200 degree to 600 degree; hence, there is high hope for implementation in industrial furnaces, cars, etc. When applied to gas-powered vehicles, 500~1,000W of electrical energy can be recycled, increasing the energy use efficiency.
If the device is applied to 30% of the cars in Japan, we can expect a 960,000kl oil substitution effect which is 1.5 times the assumed effect from solar energy generation of the year 2010. Also, if the device is implemented in industrial shaft furnaces which have an approximately 10% energy use efficiency, the efficiency will increase by 1.5 times and CO2 emission will be reduced by one-third. Iida’s research team is continuing research for further improvement in energy conversion and durability of the device.
Background of R&D and Success of Material Development;
Today, our main source of energy is fossil fuel. However the energy use efficiency has only gone up to 30% leaving 70% to be disposed as “wasted heat”. Efficiently reusing “wasted heat” and reducing fossil fuel consumption and CO2 emission, from the perspective of preventing global warming, has attracted attention from all around the world. The popularization of power generation from wasted heat has gone through many obstacles such as scarcity of material, cost, toxicity, etc. Iida’s research team has focused on the low cost and environmentally friendly silicon.
They have successfully managed to mass produce Magnesium silicide (Mg2Si), from the by-product of Si-LSI and solar cell cast wafer production, providing high-performance in power generation driven by wasted heat.
- Progress of Research -
Discovery of Magnesium silicide, material for Thermoelectric Conversion driven by Wasted Heat
Heretofore the compound of Lead and Tellurium (Pb-Te), have been known to be the material for thermoelectric conversion from wasted heat. However, Lead being hazardous and Tellurium being scarce, development of new material of low environmental burden was hoped for. Iida’s research team has been successful in being pioneers of mass producing Magnesium silicide from Silicon which is found abundantly on earth, holding the characteristic of being nonhazardous. Through this discovery of environmentally benign material, the realization of an environmentally benign technology of thermoelectric conversion through wasted heat was made possible.
Development of Magnesium silicide from Silicon by-product Silicon, being the main raw material for Magnesium silicide, is widely known to be a necessary ingredient for semiconductors throughout the electronic industry. However, in production of ultrapure silicon, much energy is consumed and more than half of the material comes out as Silicon sludge, usually being disposed of. This not only pollutes the environment, but raises the cost of production and also prevents further development of new materials and/or technology.
The research team has been successful in mass producing material for thermoelectric conversion through this waste product at a substantially low cost. Through this, thermoelectric conversion from wasted heat has become even more environmentally benign.
Profile - Tsutomu Iida
Background:
March, 1995: Meiji University Graduate School – completed PhD course
April, 1995: Japan Society for the Promotion of Science – Fellowship
July, 1995: Federal Republic of Germany – Volkswagen Foundation
April, 1997: Tokyo University of Science - Faculty of Industrial
Science and Technology – Materials Science and Technology
To Present
Major Field: Semiconductor Material Engineering
Field of Research: Semiconductor Energy Material (Thermoelectric Material / Solar Cell Material) ‘Environmentally Friendly’ Semiconductor Material
Research Content: Due to mass consumption of fossil fuel and also
for prevention of global warming, research and development of energy conversion materials are being conducted. Solar energy being the main source of reusable energy, development of solar cell material and thermoelectric conversion material is being conducted. Due to the fact that many elemental devices which are used for energy conversion tending to be toxic, development of environmentally benign semiconductor energy material continues to be conducted. Environmentally benign semiconductors are composed of semiconductor material which abundantly exists on earth and is highly ‘earth-friendly’.
Research Content:
1. Development of Thermoelectric Conversion Elemental Device through Magnesium silicide
2. Development of High Efficiency Solar Cells through Silicon Germanium
3. Photodecomposition and Hydrogen Composition of water through Semiconductor Photocatalyst
Website:http://web.mac.com/iida_lab/
Contact for this information -
Tokyo University of Science Technology Licensing Organization
Administrator: Niki
TEL: 03-5225-1089
e-mail:niki_tamotsu@admin.tus.ac.jp
If the device is applied to 30% of the cars in Japan, we can expect a 960,000kl oil substitution effect which is 1.5 times the assumed effect from solar energy generation of the year 2010.
A research team led by associate professor Tsutomu Iida (Tokyo University of Science - Faculty of Industrial Science and Technology – Materials Science and Technology) has developed a power generation unit driven by wasted heat composed of Magnesium silicide (Mg2Si), a filtered by-product of Si-LSI and solar cell cast wafer production.
This device, being the first in the world, has been successful in bulk-quantity synthesis, at the same time increasing the thermoelectric conversion rate. Approximately 2,500W/m² (per unit) of power generation and 3,000 hrs of continuous operation has been made possible, sufficiently fulfilling the criteria for commercial use.
By applying this device in industrial shaft furnaces and/or car engines, we can expect drastic reduction in fuel consumption and prevention of global warming. Implementation of this device has already been determined partially as an experiment for practical use and there are high expectations of application of this device in industrial furnaces nation-wide.
Mg2Si Power Generation Unit Driven by Waste Heat - Capability & Effect; This device has an effective temperature range from 200 degree to 600 degree; hence, there is high hope for implementation in industrial furnaces, cars, etc. When applied to gas-powered vehicles, 500~1,000W of electrical energy can be recycled, increasing the energy use efficiency.
If the device is applied to 30% of the cars in Japan, we can expect a 960,000kl oil substitution effect which is 1.5 times the assumed effect from solar energy generation of the year 2010. Also, if the device is implemented in industrial shaft furnaces which have an approximately 10% energy use efficiency, the efficiency will increase by 1.5 times and CO2 emission will be reduced by one-third. Iida’s research team is continuing research for further improvement in energy conversion and durability of the device.
Background of R&D and Success of Material Development;
Today, our main source of energy is fossil fuel. However the energy use efficiency has only gone up to 30% leaving 70% to be disposed as “wasted heat”. Efficiently reusing “wasted heat” and reducing fossil fuel consumption and CO2 emission, from the perspective of preventing global warming, has attracted attention from all around the world. The popularization of power generation from wasted heat has gone through many obstacles such as scarcity of material, cost, toxicity, etc. Iida’s research team has focused on the low cost and environmentally friendly silicon.
They have successfully managed to mass produce Magnesium silicide (Mg2Si), from the by-product of Si-LSI and solar cell cast wafer production, providing high-performance in power generation driven by wasted heat.
- Progress of Research -
Discovery of Magnesium silicide, material for Thermoelectric Conversion driven by Wasted Heat
Heretofore the compound of Lead and Tellurium (Pb-Te), have been known to be the material for thermoelectric conversion from wasted heat. However, Lead being hazardous and Tellurium being scarce, development of new material of low environmental burden was hoped for. Iida’s research team has been successful in being pioneers of mass producing Magnesium silicide from Silicon which is found abundantly on earth, holding the characteristic of being nonhazardous. Through this discovery of environmentally benign material, the realization of an environmentally benign technology of thermoelectric conversion through wasted heat was made possible.
Development of Magnesium silicide from Silicon by-product Silicon, being the main raw material for Magnesium silicide, is widely known to be a necessary ingredient for semiconductors throughout the electronic industry. However, in production of ultrapure silicon, much energy is consumed and more than half of the material comes out as Silicon sludge, usually being disposed of. This not only pollutes the environment, but raises the cost of production and also prevents further development of new materials and/or technology.
The research team has been successful in mass producing material for thermoelectric conversion through this waste product at a substantially low cost. Through this, thermoelectric conversion from wasted heat has become even more environmentally benign.
Profile - Tsutomu Iida
Background:
March, 1995: Meiji University Graduate School – completed PhD course
April, 1995: Japan Society for the Promotion of Science – Fellowship
July, 1995: Federal Republic of Germany – Volkswagen Foundation
April, 1997: Tokyo University of Science - Faculty of Industrial
Science and Technology – Materials Science and Technology
To Present
Major Field: Semiconductor Material Engineering
Field of Research: Semiconductor Energy Material (Thermoelectric Material / Solar Cell Material) ‘Environmentally Friendly’ Semiconductor Material
Research Content: Due to mass consumption of fossil fuel and also
for prevention of global warming, research and development of energy conversion materials are being conducted. Solar energy being the main source of reusable energy, development of solar cell material and thermoelectric conversion material is being conducted. Due to the fact that many elemental devices which are used for energy conversion tending to be toxic, development of environmentally benign semiconductor energy material continues to be conducted. Environmentally benign semiconductors are composed of semiconductor material which abundantly exists on earth and is highly ‘earth-friendly’.
Research Content:
1. Development of Thermoelectric Conversion Elemental Device through Magnesium silicide
2. Development of High Efficiency Solar Cells through Silicon Germanium
3. Photodecomposition and Hydrogen Composition of water through Semiconductor Photocatalyst
Website:http://web.mac.com/iida_lab/
Contact for this information -
Tokyo University of Science Technology Licensing Organization
Administrator: Niki
TEL: 03-5225-1089
e-mail:niki_tamotsu@admin.tus.ac.jp
Indus Script Encodes Language, Reveals New Study of Ancient Symbols
The Rosetta Stone allowed 19th century scholars to translate symbols left by an ancient civilization and thus decipher the meaning of Egyptian hieroglyphics.
But the symbols found on many other ancient artifacts remain a mystery, including those of a people that inhabited the Indus valley on the present-day border between Pakistan and India. Some experts question whether the symbols represent a language at all, or are merely pictograms that bear no relation to the language spoken by their creators.
A University of Washington computer scientist has led a statistical study of the Indus script, comparing the pattern of symbols to various linguistic scripts and nonlinguistic systems, including DNA and a computer programming language. The results, published online Thursday by the journal Science, found the Indus script's pattern is closer to that of spoken words, supporting the hypothesis that it codes for an as-yet-unknown language.
"We applied techniques of computer science, specifically machine learning, to an ancient problem," said Rajesh Rao, a UW associate professor of computer science and engineering and lead author of the study. "At this point we can say that the Indus script seems to have statistical regularities that are in line with natural languages."
Co-authors are Nisha Yadav and Mayank Vahia at the Tata Institute of Fundamental Research in Mumbai, India; Hrishikesh Joglekar, a software engineer from Mumbai; R. Adhikari at the Institute of Mathematical Sciences in Chennai, India; and Iravatham Mahadevan at the Indus Research Center in Chennai. The research was supported by the Packard Foundation and the Sir Jamsetji Tata Trust.
The Indus people were contemporaries of the Egyptian and Mesopotamian civilizations, inhabiting the Indus river valley in present-day eastern Pakistan and northwestern India from about 2600 to 1900 B.C. This was an advanced, urbanized civilization that left written symbols on tiny stamp seals, amulets, ceramic objects and small tablets.
"The Indus script has been known for almost 130 years," said Rao, an Indian native with a longtime personal interest in the subject. "Despite more than 100 attempts, it has not yet been deciphered. The underlying assumption has always been that the script encodes language."
In 2004 a provocative paper titled The Collapse of the Indus-Script Thesis claimed that the short inscriptions have no linguistic content and are merely brief pictograms depicting religious or political symbols. That paper's lead author offered a $10,000 reward to anybody who could produce an Indus artifact with more than 50 symbols.
Taking a scientific approach, the U.S.-Indian team of computer scientists and mathematicians looked at the statistical patterns in sequences of Indus symbols. They calculated the amount of randomness allowed in choosing the next symbol in a sequence. Some nonlinguistic systems display a random pattern, while others, such as pictures that represent deities, follow a strict order that reflects some underlying hierarchy. Spoken languages tend to fall between the two extremes, incorporating some order as well as some flexibility.
The new study compared a well-known compilation of Indus texts with linguistic and nonlinguistic samples. The researchers performed calculations on present-day texts of English; texts of the Sumerian language spoken in Mesopotamia during the time of the Indus civilization; texts in Old Tamil, a Dravidian language originating in southern India that some scholars have hypothesized is related to the Indus script; and ancient Sanskrit, one of the earliest members of the Indo-European language family. In each case the authors calculated the conditional entropy, or randomness, of the symbols' order.
They then repeated the calculations for samples of symbols that are not spoken languages: one in which the placement of symbols was completely random; another in which the placement of symbols followed a strict hierarchy; DNA sequences from the human genome; bacterial protein sequences; and an artificially created linguistic system, the computer programming language Fortran.
Results showed that the Indus inscriptions fell in the middle of the spoken languages and differed from any of the nonlinguistic systems.
If the Indus symbols are a spoken language, then deciphering them would open a window onto a civilization that lived more than 4,000 years ago. The researchers hope to continue their international collaboration, using a mathematical approach to delve further into the Indus script.
"We would like to make as much headway as possible and ideally, yes, we'd like to crack the code," Rao said. "For now we want to analyze the structure and syntax of the script and infer its grammatical rules. Someday we could leverage this information to get to a decipherment, if, for example, an Indus equivalent of the Rosetta Stone is unearthed in the future."
More information about the Indus civilization and language is at http://www.harappa.com
The Rosetta Stone allowed 19th century scholars to translate symbols left by an ancient civilization and thus decipher the meaning of Egyptian hieroglyphics.
But the symbols found on many other ancient artifacts remain a mystery, including those of a people that inhabited the Indus valley on the present-day border between Pakistan and India. Some experts question whether the symbols represent a language at all, or are merely pictograms that bear no relation to the language spoken by their creators.
A University of Washington computer scientist has led a statistical study of the Indus script, comparing the pattern of symbols to various linguistic scripts and nonlinguistic systems, including DNA and a computer programming language. The results, published online Thursday by the journal Science, found the Indus script's pattern is closer to that of spoken words, supporting the hypothesis that it codes for an as-yet-unknown language.
"We applied techniques of computer science, specifically machine learning, to an ancient problem," said Rajesh Rao, a UW associate professor of computer science and engineering and lead author of the study. "At this point we can say that the Indus script seems to have statistical regularities that are in line with natural languages."
Co-authors are Nisha Yadav and Mayank Vahia at the Tata Institute of Fundamental Research in Mumbai, India; Hrishikesh Joglekar, a software engineer from Mumbai; R. Adhikari at the Institute of Mathematical Sciences in Chennai, India; and Iravatham Mahadevan at the Indus Research Center in Chennai. The research was supported by the Packard Foundation and the Sir Jamsetji Tata Trust.
The Indus people were contemporaries of the Egyptian and Mesopotamian civilizations, inhabiting the Indus river valley in present-day eastern Pakistan and northwestern India from about 2600 to 1900 B.C. This was an advanced, urbanized civilization that left written symbols on tiny stamp seals, amulets, ceramic objects and small tablets.
"The Indus script has been known for almost 130 years," said Rao, an Indian native with a longtime personal interest in the subject. "Despite more than 100 attempts, it has not yet been deciphered. The underlying assumption has always been that the script encodes language."
In 2004 a provocative paper titled The Collapse of the Indus-Script Thesis claimed that the short inscriptions have no linguistic content and are merely brief pictograms depicting religious or political symbols. That paper's lead author offered a $10,000 reward to anybody who could produce an Indus artifact with more than 50 symbols.
Taking a scientific approach, the U.S.-Indian team of computer scientists and mathematicians looked at the statistical patterns in sequences of Indus symbols. They calculated the amount of randomness allowed in choosing the next symbol in a sequence. Some nonlinguistic systems display a random pattern, while others, such as pictures that represent deities, follow a strict order that reflects some underlying hierarchy. Spoken languages tend to fall between the two extremes, incorporating some order as well as some flexibility.
The new study compared a well-known compilation of Indus texts with linguistic and nonlinguistic samples. The researchers performed calculations on present-day texts of English; texts of the Sumerian language spoken in Mesopotamia during the time of the Indus civilization; texts in Old Tamil, a Dravidian language originating in southern India that some scholars have hypothesized is related to the Indus script; and ancient Sanskrit, one of the earliest members of the Indo-European language family. In each case the authors calculated the conditional entropy, or randomness, of the symbols' order.
They then repeated the calculations for samples of symbols that are not spoken languages: one in which the placement of symbols was completely random; another in which the placement of symbols followed a strict hierarchy; DNA sequences from the human genome; bacterial protein sequences; and an artificially created linguistic system, the computer programming language Fortran.
Results showed that the Indus inscriptions fell in the middle of the spoken languages and differed from any of the nonlinguistic systems.
If the Indus symbols are a spoken language, then deciphering them would open a window onto a civilization that lived more than 4,000 years ago. The researchers hope to continue their international collaboration, using a mathematical approach to delve further into the Indus script.
"We would like to make as much headway as possible and ideally, yes, we'd like to crack the code," Rao said. "For now we want to analyze the structure and syntax of the script and infer its grammatical rules. Someday we could leverage this information to get to a decipherment, if, for example, an Indus equivalent of the Rosetta Stone is unearthed in the future."
More information about the Indus civilization and language is at http://www.harappa.com
Thursday, April 23, 2009
New Family of Proteins - TPC2
International research collaborators have identified a new family of proteins, TPC2 (two-pore channels), that facilitates calcium signaling from specialized subcellular organelles. It is the first to isolate TPC2 as a channel that binds to nucleotide nicotinic acid adenine dinucleotide phosphate (NAADP), a second-signaling messenger, resulting in the release of calcium from intracellular stores. According to the researchers, this new discovery may have broad implications in cell biology and human disease research.
“The discovery was the result of many researchers working as one international team toward a unified outcome. We are very appreciative of all the collaborators’ efforts,” said Jianjie Ma, PhD, professor of physiology and biophysics at UMDNJ-Robert Wood Johnson Medical School. “We are proud to be part of a study that will stand as the foundation for further exploration of human disease, helping researchers to better understand how calcium contributes to cell growth and disorders, including aging-related cardiac disease, diabetes, lysosomal cell dysfunction and the metastasis of cells in cancer.”
According to the researchers, the mechanism for how NAADP triggers the release of calcium, as well as the specific sites of calcium store targeted for release, were previously unknown. These findings indicate that NAADP, through its interaction with TPC2, targets a specific store of calcium in lysosomes, a specialized subunit within the cell that contain digestion enzymes and regulate cell function.
The study was a collaboration of investigative teams at four universities, including the laboratory of Dr. Michael Zhu at the Ohio State University, the laboratory of Dr. A. Mark Evans at the University of Edinburgh and the laboratory of Dr. Antony Galione at the University of Oxford.
The research was supported by grants from the United Kingdom’s Wellcome Trust and the British Heart Foundation, the United States’ National Institutes of Health, and the American Heart Association.
UMDNJ-ROBERT WOOD JOHNSON MEDICAL SCHOOL
As one of the nation’s leading comprehensive medical schools, Robert Wood Johnson Medical School of the University of Medicine and Dentistry of New Jersey is dedicated to the pursuit of excellence in education, research, health care delivery, and the promotion of community health. In cooperation with Robert Wood Johnson University Hospital, the medical school’s principal affiliate, they comprise New Jersey’s premier academic medical center. In addition, Robert Wood Johnson Medical School has 34 hospital affiliates and ambulatory care sites throughout the region.
As one of the eight schools of the University of Medicine and Dentistry of New Jersey with 2,800 full-time and volunteer faculty, Robert Wood Johnson Medical School encompasses 22 basic science and clinical departments and hosts centers and institutes including The Cancer Institute of New Jersey, the Child Health Institute of New Jersey, the Center for Advanced Biotechnology and Medicine, the Environmental and Occupational Health Sciences Institute, and the Stem Cell Institute of New Jersey. The medical school maintains educational programs at the undergraduate, graduate and postgraduate levels for more than 1,500 students on its campuses in New Brunswick, Piscataway, and Camden, and provides continuing education courses for health care professionals and community education programs.
International research collaborators have identified a new family of proteins, TPC2 (two-pore channels), that facilitates calcium signaling from specialized subcellular organelles. It is the first to isolate TPC2 as a channel that binds to nucleotide nicotinic acid adenine dinucleotide phosphate (NAADP), a second-signaling messenger, resulting in the release of calcium from intracellular stores. According to the researchers, this new discovery may have broad implications in cell biology and human disease research.
“The discovery was the result of many researchers working as one international team toward a unified outcome. We are very appreciative of all the collaborators’ efforts,” said Jianjie Ma, PhD, professor of physiology and biophysics at UMDNJ-Robert Wood Johnson Medical School. “We are proud to be part of a study that will stand as the foundation for further exploration of human disease, helping researchers to better understand how calcium contributes to cell growth and disorders, including aging-related cardiac disease, diabetes, lysosomal cell dysfunction and the metastasis of cells in cancer.”
According to the researchers, the mechanism for how NAADP triggers the release of calcium, as well as the specific sites of calcium store targeted for release, were previously unknown. These findings indicate that NAADP, through its interaction with TPC2, targets a specific store of calcium in lysosomes, a specialized subunit within the cell that contain digestion enzymes and regulate cell function.
The study was a collaboration of investigative teams at four universities, including the laboratory of Dr. Michael Zhu at the Ohio State University, the laboratory of Dr. A. Mark Evans at the University of Edinburgh and the laboratory of Dr. Antony Galione at the University of Oxford.
The research was supported by grants from the United Kingdom’s Wellcome Trust and the British Heart Foundation, the United States’ National Institutes of Health, and the American Heart Association.
UMDNJ-ROBERT WOOD JOHNSON MEDICAL SCHOOL
As one of the nation’s leading comprehensive medical schools, Robert Wood Johnson Medical School of the University of Medicine and Dentistry of New Jersey is dedicated to the pursuit of excellence in education, research, health care delivery, and the promotion of community health. In cooperation with Robert Wood Johnson University Hospital, the medical school’s principal affiliate, they comprise New Jersey’s premier academic medical center. In addition, Robert Wood Johnson Medical School has 34 hospital affiliates and ambulatory care sites throughout the region.
As one of the eight schools of the University of Medicine and Dentistry of New Jersey with 2,800 full-time and volunteer faculty, Robert Wood Johnson Medical School encompasses 22 basic science and clinical departments and hosts centers and institutes including The Cancer Institute of New Jersey, the Child Health Institute of New Jersey, the Center for Advanced Biotechnology and Medicine, the Environmental and Occupational Health Sciences Institute, and the Stem Cell Institute of New Jersey. The medical school maintains educational programs at the undergraduate, graduate and postgraduate levels for more than 1,500 students on its campuses in New Brunswick, Piscataway, and Camden, and provides continuing education courses for health care professionals and community education programs.
Monday, April 20, 2009
How genes are controlled in mammals
The international FANTOM consortium announces publication of three milestone papers in the prestigious journal Nature Genetics that will challenge current notions of how genes are controlled in mammals.
FANTOM, or Functional Annotation of the Mammalian cDNA, which is organized by RIKEN Omics Science Center (OSC), has leading scientists in Australia, Switzerland, Norway, South Africa, Sweden, Canada, Denmark, Italy, Germany, Singapore, UK, and the United States. The consortium has been providing the scientific community with extensive databases on the mammalian genome that describe molecular function, biology, and cell components. FANTOM has become a world authority on the mammalian transcriptome, the set of all messenger RNA showing active genetic expression at one point in time. Other major discoveries are that approximately 70% of the genome is transcribed and that more than half of the expressed genes are likely non-coding RNAs (ncRNAs) that do not code proteins; thus, the prevailing theory that only 2% of the genome is transcribed into mRNA coding to proteins needed to be reexamined. Now in its fourth stage, FANTOM4, led by OSC’s Dr. Yoshihide Hayashizaki, has in over 3 years of laborious research developed a novel technology for producing a genome-wide promoter expression profile, established a mathematical scheme for describing the data obtained, and extracted key genomic elements that play dominant roles in the maintenance of cellular conditions.
In the current research, OSC has broadened its original technology CAGE (Cap Analysis of Gene Expression) and created deepCAGE, which takes advantage of next-generation sequencing to both precisely identify transcription start sites genome wide as well as to quantify the expression of each start site. The deepCAGE technology was applied to a differentiating acute myeloid leukemia cell line (ACL) to provide genome-wide time course dynamics of expression at the level of individual promoters — specific sequences on the DNA providing binding sites for RNA polymerase and the protein transcription factors that recruit them. The consortium built a quantitative model of the genome-wide gene expression dynamics that identified the key regulator motifs driving the differentiation, the time-dependent activities of the transcription regulators binding the motifs, and the genome-wide target promoters of each motif.
Validation of the model was performed by knocking down each transcription factor with small interfering RNAs. This first report of a large-scale gene network based on experimental data set is certain to generate much excitement in the scientific community. This information is also important for life science and medical researchers who are trying to uncover the processes by which cells undergo conversion or become cancerous, and for those attempting to determine how to control the growth and differentiation of stem cells and ensure their safety for use in regenerative medicine. Dr. Harukazu Suzuki, the scientific coordinator of the consortium, had this to say, “We are proud that we have created groundbreaking research in understanding more about how genes regulate cells at the molecular level and we want to acknowledge all consortium members for their great contribution to the research effort.”
The FANTOM consortium has also expanded earlier discoveries of transcriptional complexity by exploring repetitive elements found throughout mammalian genomes with DeepCAGE. These elements, which constitute up to half of the genome, have been generally considered to be junk or parasitic DNA. However, the team has found that the repetitive elements are broadly expressed and 6 to 30% of mouse and human mRNAs are derived from repetitive element promoters. These RNAs are often tissue-specific and dynamically controlled, and control the output of the genome through a variety of mechanisms. The FANTOM4 collaborators have also identified yet another type of short RNA, referred to as tiRNA (transcription initiation RNA) or tiny RNAs, in the human, chicken, and Drosphilia. They are about 18 nucleotides (nt) in length and are found within -60 to +120 nt of transcription start sites and may actually be widespread in metazoans (animals). A BioMed Central Thematic Series features even more FANTOM 4 research papers in Genome Biology and several BMC journals.
Contact:
RIKEN Omics Science Center
Director: Yoshihide Hayashizaki
Project director: Harukazu Suzuki
TEL: +81-45-503-2222 FAX: +81-45-503-9216
The international FANTOM consortium announces publication of three milestone papers in the prestigious journal Nature Genetics that will challenge current notions of how genes are controlled in mammals.
FANTOM, or Functional Annotation of the Mammalian cDNA, which is organized by RIKEN Omics Science Center (OSC), has leading scientists in Australia, Switzerland, Norway, South Africa, Sweden, Canada, Denmark, Italy, Germany, Singapore, UK, and the United States. The consortium has been providing the scientific community with extensive databases on the mammalian genome that describe molecular function, biology, and cell components. FANTOM has become a world authority on the mammalian transcriptome, the set of all messenger RNA showing active genetic expression at one point in time. Other major discoveries are that approximately 70% of the genome is transcribed and that more than half of the expressed genes are likely non-coding RNAs (ncRNAs) that do not code proteins; thus, the prevailing theory that only 2% of the genome is transcribed into mRNA coding to proteins needed to be reexamined. Now in its fourth stage, FANTOM4, led by OSC’s Dr. Yoshihide Hayashizaki, has in over 3 years of laborious research developed a novel technology for producing a genome-wide promoter expression profile, established a mathematical scheme for describing the data obtained, and extracted key genomic elements that play dominant roles in the maintenance of cellular conditions.
In the current research, OSC has broadened its original technology CAGE (Cap Analysis of Gene Expression) and created deepCAGE, which takes advantage of next-generation sequencing to both precisely identify transcription start sites genome wide as well as to quantify the expression of each start site. The deepCAGE technology was applied to a differentiating acute myeloid leukemia cell line (ACL) to provide genome-wide time course dynamics of expression at the level of individual promoters — specific sequences on the DNA providing binding sites for RNA polymerase and the protein transcription factors that recruit them. The consortium built a quantitative model of the genome-wide gene expression dynamics that identified the key regulator motifs driving the differentiation, the time-dependent activities of the transcription regulators binding the motifs, and the genome-wide target promoters of each motif.
Validation of the model was performed by knocking down each transcription factor with small interfering RNAs. This first report of a large-scale gene network based on experimental data set is certain to generate much excitement in the scientific community. This information is also important for life science and medical researchers who are trying to uncover the processes by which cells undergo conversion or become cancerous, and for those attempting to determine how to control the growth and differentiation of stem cells and ensure their safety for use in regenerative medicine. Dr. Harukazu Suzuki, the scientific coordinator of the consortium, had this to say, “We are proud that we have created groundbreaking research in understanding more about how genes regulate cells at the molecular level and we want to acknowledge all consortium members for their great contribution to the research effort.”
The FANTOM consortium has also expanded earlier discoveries of transcriptional complexity by exploring repetitive elements found throughout mammalian genomes with DeepCAGE. These elements, which constitute up to half of the genome, have been generally considered to be junk or parasitic DNA. However, the team has found that the repetitive elements are broadly expressed and 6 to 30% of mouse and human mRNAs are derived from repetitive element promoters. These RNAs are often tissue-specific and dynamically controlled, and control the output of the genome through a variety of mechanisms. The FANTOM4 collaborators have also identified yet another type of short RNA, referred to as tiRNA (transcription initiation RNA) or tiny RNAs, in the human, chicken, and Drosphilia. They are about 18 nucleotides (nt) in length and are found within -60 to +120 nt of transcription start sites and may actually be widespread in metazoans (animals). A BioMed Central Thematic Series features even more FANTOM 4 research papers in Genome Biology and several BMC journals.
Contact:
RIKEN Omics Science Center
Director: Yoshihide Hayashizaki
Project director: Harukazu Suzuki
TEL: +81-45-503-2222 FAX: +81-45-503-9216
Wednesday, April 01, 2009
Distinguishing Single Cells With Nothing But Light
Researchers at the University of Rochester have developed a novel optical technique that permits rapid analysis of single human immune cells using only light.
Availability of such a technique means that immunologists and other cellular researchers may soon be able to observe the responses of individual cells to various stimuli, rather than relying on aggregate statistical data from large cell populations. Until now scientists have not had a non-invasive way to see how human cells, like T cells or cancer cells, activate individually and evolve over time.
As reported today in a special biomedical issue of Applied Optics, this is the first time clear differences between two types of immune cells have been seen using a microscopy system that gathers chemical and structural information by combining two previously distinct optical techniques, according to senior author Andrew Berger, associate professor of optics at the University of Rochester.
Berger and his graduate student Zachary Smith are the first to integrate Raman and angular-scattering microscopy into a single system, which they call IRAM.
“Conceptually it’s pretty straightforward—you shine a specified wavelength of light onto your sample and you get back a large number of peaks spread out like a rainbow,” says Berger. “The peaks tell you how the molecules you’re studying vibrate and together the vibrations give you the chemical information.”
According to Smith, “Raman spectroscopy is essentially an easy way to get a fingerprint from the molecule.”
Structural information is simultaneously gathered by examining the angles at which light incident on a sample is bumped off its original course.
Together the chemical and structural information provide the data needed to classify and distinguish between two different, single cells. Berger and Smith verified this by looking at single granulocytes—a type of white blood cell—and peripheral blood monocytes.
“One of the big plusses with our system is that it’s a non-labeling approach for studying living cells,” says Berger.
IRAM differs from most standard procedures where markers are inserted in, or attached to cells. If a marker sticks to one cell, and not the other, you can tell which cell is which on the basis of specific binding properties.
While markers are often adequate for studying cells at a single point in time, monitoring a cell over time as it changes is more problematic, since the marker can affect dynamic cell activities, like membrane transport. And internal markers actually involve punching holes in the membrane, damaging or killing the cell in the process.
“Our method uses only light to effectively reach inside the cell,” says Smith. “We can classify internal differences in the cell without opening it up, attaching anything to it, or preparing it in any special way. It’s really just flipping a switch.”
Despite being relatively intense, the light used with IRAM does not harm or inhibit normal cell functionality. This is because the wavelength of the light can be precisely calibrated to minimize absorption by the cells. The near-infrared spectrum has proven particularly optimal for allowing almost all of the light to pass through the cells.
With the availability of a technique where making a measurement does not alter cellular activity, scientists will be able to better observe individual cell responses to stimuli, which Berger and Smith suspect may have far reaching implications for current understandings of cell activation and development.
“In the cell sensing community it’s currently a pretty hot area to figure out how to analyze activation responses on a cell-by-cell basis,” says Berger. “If individual information was available on top of existing ensemble data, you’d have a richer understanding of immune responses.”
Perfecting IRAM has been a stepping stone process so far. Now that individual cells can be distinguished, Berger and Smith are actively investigating activation processes more explicitly. Preliminary IRAM experiments conducted on T cells have revealed perceivable differences between the initial resting state of a T cell and its state following an encounter with an invader.
The next step will be to use IRAM to gather data continuously so that scientists can effectively watch single cells undergo activation and react to stimuli in real-time. The ability to know not only about the aggregate responses of cells, but also be able to observe the earliest changes among individual cells, may be of profound importance in time-critical areas, such as cancer research and immunology.
“There’s an obvious desire among cell researchers to be able to deliver a controlled stimulant to a single cell and then study its response over time,” says Berger. “The clinical insights that might arise are currently in the realm of speculation. We won’t know until we can do it—and now we can.”
Researchers at the University of Rochester have developed a novel optical technique that permits rapid analysis of single human immune cells using only light.
Availability of such a technique means that immunologists and other cellular researchers may soon be able to observe the responses of individual cells to various stimuli, rather than relying on aggregate statistical data from large cell populations. Until now scientists have not had a non-invasive way to see how human cells, like T cells or cancer cells, activate individually and evolve over time.
As reported today in a special biomedical issue of Applied Optics, this is the first time clear differences between two types of immune cells have been seen using a microscopy system that gathers chemical and structural information by combining two previously distinct optical techniques, according to senior author Andrew Berger, associate professor of optics at the University of Rochester.
Berger and his graduate student Zachary Smith are the first to integrate Raman and angular-scattering microscopy into a single system, which they call IRAM.
“Conceptually it’s pretty straightforward—you shine a specified wavelength of light onto your sample and you get back a large number of peaks spread out like a rainbow,” says Berger. “The peaks tell you how the molecules you’re studying vibrate and together the vibrations give you the chemical information.”
According to Smith, “Raman spectroscopy is essentially an easy way to get a fingerprint from the molecule.”
Structural information is simultaneously gathered by examining the angles at which light incident on a sample is bumped off its original course.
Together the chemical and structural information provide the data needed to classify and distinguish between two different, single cells. Berger and Smith verified this by looking at single granulocytes—a type of white blood cell—and peripheral blood monocytes.
“One of the big plusses with our system is that it’s a non-labeling approach for studying living cells,” says Berger.
IRAM differs from most standard procedures where markers are inserted in, or attached to cells. If a marker sticks to one cell, and not the other, you can tell which cell is which on the basis of specific binding properties.
While markers are often adequate for studying cells at a single point in time, monitoring a cell over time as it changes is more problematic, since the marker can affect dynamic cell activities, like membrane transport. And internal markers actually involve punching holes in the membrane, damaging or killing the cell in the process.
“Our method uses only light to effectively reach inside the cell,” says Smith. “We can classify internal differences in the cell without opening it up, attaching anything to it, or preparing it in any special way. It’s really just flipping a switch.”
Despite being relatively intense, the light used with IRAM does not harm or inhibit normal cell functionality. This is because the wavelength of the light can be precisely calibrated to minimize absorption by the cells. The near-infrared spectrum has proven particularly optimal for allowing almost all of the light to pass through the cells.
With the availability of a technique where making a measurement does not alter cellular activity, scientists will be able to better observe individual cell responses to stimuli, which Berger and Smith suspect may have far reaching implications for current understandings of cell activation and development.
“In the cell sensing community it’s currently a pretty hot area to figure out how to analyze activation responses on a cell-by-cell basis,” says Berger. “If individual information was available on top of existing ensemble data, you’d have a richer understanding of immune responses.”
Perfecting IRAM has been a stepping stone process so far. Now that individual cells can be distinguished, Berger and Smith are actively investigating activation processes more explicitly. Preliminary IRAM experiments conducted on T cells have revealed perceivable differences between the initial resting state of a T cell and its state following an encounter with an invader.
The next step will be to use IRAM to gather data continuously so that scientists can effectively watch single cells undergo activation and react to stimuli in real-time. The ability to know not only about the aggregate responses of cells, but also be able to observe the earliest changes among individual cells, may be of profound importance in time-critical areas, such as cancer research and immunology.
“There’s an obvious desire among cell researchers to be able to deliver a controlled stimulant to a single cell and then study its response over time,” says Berger. “The clinical insights that might arise are currently in the realm of speculation. We won’t know until we can do it—and now we can.”
Subscribe to:
Posts (Atom)
